The draft of my final project, due 10 Nov. After I received feedback on my outline, I decided to remove a lot of the treatment groups, and instead chose to focus on one specific miRNA. I was very proud of how far that I’d come since the question, and even since the outline!
Final Project Outline
The outline of my final project, due 25 October. It’s interesting to follow the development from question to outline, and how I added more!
DocuLearn 4
1. I think that the most challenging aspect was getting all of my thoughts in order. I would keep thinking up ideas for the experiment, some practical and some not, and I would have to figure out how to expand on that idea or whether or not that idea would even work for its intended purpose. To overcome this challenge I made sure to write down any and all ideas that I had when I had them, and then search and see if any other papers had used similar techniques to perform experiments. Once I got the ideas down on paper, it was a lot easier to see which ones were practical and which ones wouldn’t work. I think that by writing them down I actually had to think through the steps of that particular process, and how it may apply to my project, which helped me figure out both more about my project and, through researching, more about the techniques I was thinking of.
2. I think that I have learned that I am capable of thinking up an experiment! Before, I would read all of these papers and just wonder how on earth they thought up that idea, or came up with that experiment to test some question. Now I see that I can come up with questions, and find resources that help provide background. I have also improved my knowledge of various techniques. By having to find which ones are applicable to my project I have had to read a lot more in depth to various techniques, allowing me to become much more comfortable with them as well as understand when and how they may be used. I am surprised by how much I enjoyed researching this project, and how exciting it was to think of experiments and background information that I would need to know. It made learning fun, to be working on something I enjoyed. This project has really shown me that research is what I want to do in the future, and that I do have the skills and creativity necessary to pursue that.
Case Study Stage 5 – Summary
Honey Bee Caste Differentiation: Case Study pt. 5:
i) The honeybee paper that we analyzed was the paper titled “Royalactin induces queen differentiation in honeybees” by Masaki Kamakura. A full reference is below:
Kamakura M (2011) Royalactin induces queen differentiation in honeybees. Nature 473: 478–483.
A copy of my completed worksheet is also available below:
ii) The mechanistic model that my group developed is seen below:
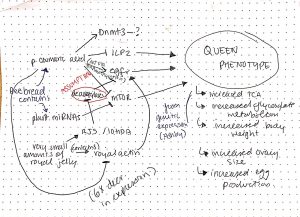
Figure 1: This model focuses on broader inhibitory and activating effects between various proteins that may lead to the queen phenotype. The exact nature of any biochemical interaction is not known. The arrows in the diagram are all strongly supported by articles in class as well as some outside sources, except for that connecting 10HDA to egfr upregulation, as that is an assumption (10HDA is a HDACi, and could theoretically act in any of the pathways in the diagram to carry out its effect, but we theorize here that it acts on egfr due to phenotypic effects associated with egfr).
Group members: Heather Betz (me), Erin Yang, Hayden Wood, Jamie Vanden Broek, Maria Saleeb
iii) There was no copy that the class converged to, but ours seemed to encompass the most outside information in its development (including articles that contradicted Kamakura 2011), so for this I would use our model.
iv)
- Summary of model: Regulation of the Queen phenotype is a complex process, with the major proteins leading to the development of the phenotype being egfr, ILP2, and mTOR, with other proteins such as p. coumaric acid and royalactin functioning to regulate the production of those proteins. On a broad scale, p. coumaric acid and plant miRNAs from beebread work to suppress Queen phenotype development, whereas royalactin and 10HDA from royal jelly activate its development.
- Contradictory evidence: Kamakura said that royalactin was responsible for queen phenotype expression through upregulation of egfr, but other papers such as Buttstedt et. al found this not to be the case, and therefore this should be clarified through further experiments whether or not royalactin is solely responsible. Kucharski et. al also argued that EGFR gene methylation (which Kamakura hypothesized as occurring by royalactin) was not playing a role in phenotype expression, which should also be clarified.
- Suggestions: One experiment, the most crucial, should determine whether or not the royalactin in royal jelly is responsible for the upregulation of egfr, which in turn encourages the Queen phenotype. Another experiment would be to determine how 10HDA fits in to the process, and what exactly it’s acting on to help with caste differentiation.
v) References:
- Ashby et. al (2016) MicroRNAs in honey bee caste determination. Scientific Reports 6, 18794 (online)
- Buttstedt et. al (2016) Royalactin is not a royal making of a queen. Nature 537: E10-E12
- Kamakura M (2011) Royalactin induces queen differentiation in honeybees. Nature 473: 478–483.
- Kucharski et. al (2015) EGFR gene methylation is not involved in Royalactin controlled phenotypic polymorphism in honey bees. Scientific Reports 5: ISSN 2045-2322 (online).
- Mao et. al (2015). A dietary phytochemical alters caste-associated gene expression in honey bees. Science Advances 1, e1500795 (online).
- Spannhoff et. al (2011) Histone deacetylase inhibitor activity in royal jelly might facilitate caste switching in bees. EMBO reports 12, 238-243
- Zhu et. al (2017). Plant microRNAs in larval food regulate honeybee caste development. PLOS Genetics 13, e1006946 (online).
DocuLearn 3
- I have learned several things through this unit, many of them being specific to honeybees and their caste differentiation, such as how EGFR plays a role. Several things I learned are more broadly applicable, such as how there can be many different explanations for the same phenomenon, and how authors can reply to papers published if they have data that shows differing outcomes.
- I have been practicing parsing relevant information from many different sources, and using that information to create models that incorporate ideas from different sources to display a more cohesive approach to an idea. This would be very useful to have both in and outside of this class.
- I find that my challenges in persisting through reading papers that I described in the first DocuLearn are the challenges that I had in this assignment, rather than issues with understanding techniques used, as I have started to just look up any technique that I come across that I am not familiar with.
Final Project Potential Question
Can the 10 miRNAs that are common to and present in regeneration sites in species that are capable of limb regeneration (zebrafish, axolotls…) aid in healing of human cutaneous wounds such as lacerations and incisions?
DocuLearn 2
1. Two things that I’ve learned are as follows: I’ve learned about the concept of self-regulation and have found myself being more conscious about learning for me, rather than because I feel like I have to. I’ve also learned about the difference between specialization and determination, and how these are both different from differentiation (I used to think that they were all pretty much the same thing.) Of course, I’ve learned more than this as well.
2. The second thing I learned fits into my wanting to learn about how cells can be so specially adapted for a certain task. Knowing the difference between these terms is a very beginning bit to learning how complex organisms can arise, as these terms are fundamental vocabulary that I needed to understand before going forwards.
3. I think the most challenging aspect of the course is discussions, and coming up with experiments to investigate questions. I find that I can think up broad ideas for what I would want to accomplish in a potential experiment, but I get stuck when having to think up specific techniques to do so. I will have to review techniques such as CHiP-seq going forward. The most surprising aspect of the course is how much I look forward to it! I feel like I really enjoy learning about epigenetics, and I didn’t expect to find it so interesting.
DocuLearn 1
1. Two things that I would like to learn in this course are:
1) How to be discerning in reading scientific papers, and select the parts that are needed without getting too bogged down in information that I may not understand so well or is not relevant to what I’m trying to learn from the paper
2) Learn how a single cell/zygote can eventually develop into an organism that comprises millions of cells, and how each of these cells can be so specifically adapted to a purpose while still working as part of the whole.
2. For learning goal 1, I think that persisting when tasks are difficult will be the most challenging in this situation, as when I’m reading higher-level things I tend to get frustrated when I can’t understand what it’s saying easily. I will respond to this challenge by really focusing on what I need to take out of the paper, and if I get to a difficult piece that I can’t seem to understand I will take a break for a little and come back to it, and if I”m still having trouble I will try to get help from either friends in the course or my professor.
3. I think this is self-regulated learning, as it involves me specifically thinking of two things that I want to learn, and is the beginning of me setting goals that I would be self-regulated in achieving. These two things are specific to me and may not coincide with what others want to learn, and in that sense thinking of them is a very self-oriented process. Answering these questions also made me think of a way that I would persist when encountering difficulties, giving me strategies for further learning adventures.
If I Were a Developmental Biologist
1. If I were a researcher in developmental genetics/genomics, two thing that I would investigate would be:
1) What goes wrong in development of some blood cells that can lead to the Philadelphia chromosome forming during cell replication, and can this be prevented?
2) Could miRNA from species capable of regeneration (axolotl, zebrafish) that seem to control gene expression in the regeneration process be useful in wound healing/tissue regeneration in humans?
2. The potential impact of the second investigation would be paramount if the miRNA could in fact assist in wound healing, or human regeneration. Axolotls can infinitely regrow limbs, and it’s been found that there are 10 microRNAs that they have in common with with other species capable of regeneration that are all active in the same ways during limb/fin regeneration. Humans could find a way to heal injuries more naturally and fully than before if these miRNA, or their human equivalents could be found and activated, and it would open up a whole new field in gene therapy research involving regeneration.
For Assignments:
I wanted to include this because it shows how far I’ve come throughout this course, when taken in context with my final project. It shows how I was able to take something I had a casual interest in, and turn it into a complete final project, further developing it as I went. From this assignment, I learned what type of questions are being asked in developmental biology, and by looking back at it now I really know how far I have come throughout this semester.