Honey Bee Caste Differentiation: Case Study pt. 5:
i) The honeybee paper that we analyzed was the paper titled “Royalactin induces queen differentiation in honeybees” by Masaki Kamakura. A full reference is below:
Kamakura M (2011) Royalactin induces queen differentiation in honeybees. Nature 473: 478–483.
A copy of my completed worksheet is also available below:
Kamakura Royalactin Worksheet
ii) The mechanistic model that my group developed is seen below:
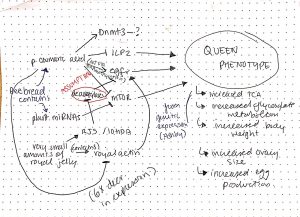
Figure 1: This model focuses on broader inhibitory and activating effects between various proteins that may lead to the queen phenotype. The exact nature of any biochemical interaction is not known. The arrows in the diagram are all strongly supported by articles in class as well as some outside sources, except for that connecting 10HDA to egfr upregulation, as that is an assumption (10HDA is a HDACi, and could theoretically act in any of the pathways in the diagram to carry out its effect, but we theorize here that it acts on egfr due to phenotypic effects associated with egfr).
Group members: Heather Betz (me), Erin Yang, Hayden Wood, Jamie Vanden Broek, Maria Saleeb
iii) There was no copy that the class converged to, but ours seemed to encompass the most outside information in its development (including articles that contradicted Kamakura 2011), so for this I would use our model.
iv)
- Summary of model: Regulation of the Queen phenotype is a complex process, with the major proteins leading to the development of the phenotype being egfr, ILP2, and mTOR, with other proteins such as p. coumaric acid and royalactin functioning to regulate the production of those proteins. On a broad scale, p. coumaric acid and plant miRNAs from beebread work to suppress Queen phenotype development, whereas royalactin and 10HDA from royal jelly activate its development.
- Contradictory evidence: Kamakura said that royalactin was responsible for queen phenotype expression through upregulation of egfr, but other papers such as Buttstedt et. al found this not to be the case, and therefore this should be clarified through further experiments whether or not royalactin is solely responsible. Kucharski et. al also argued that EGFR gene methylation (which Kamakura hypothesized as occurring by royalactin) was not playing a role in phenotype expression, which should also be clarified.
- Suggestions: One experiment, the most crucial, should determine whether or not the royalactin in royal jelly is responsible for the upregulation of egfr, which in turn encourages the Queen phenotype. Another experiment would be to determine how 10HDA fits in to the process, and what exactly it’s acting on to help with caste differentiation.
v) References:
- Ashby et. al (2016) MicroRNAs in honey bee caste determination. Scientific Reports 6, 18794 (online)
- Buttstedt et. al (2016) Royalactin is not a royal making of a queen. Nature 537: E10-E12
- Kamakura M (2011) Royalactin induces queen differentiation in honeybees. Nature 473: 478–483.
- Kucharski et. al (2015) EGFR gene methylation is not involved in Royalactin controlled phenotypic polymorphism in honey bees. Scientific Reports 5: ISSN 2045-2322 (online).
- Mao et. al (2015). A dietary phytochemical alters caste-associated gene expression in honey bees. Science Advances 1, e1500795 (online).
- Spannhoff et. al (2011) Histone deacetylase inhibitor activity in royal jelly might facilitate caste switching in bees. EMBO reports 12, 238-243
- Zhu et. al (2017). Plant microRNAs in larval food regulate honeybee caste development. PLOS Genetics 13, e1006946 (online).