TILLING Technique
(Modified from Till BJ, et al. 2003, Genome Res. 13:524-530)
The general procedure is as follows:
A population is mutagenised with EMS (or another mutagen), and allowed to grow up and produce progeny.
DNA is isolated from the progeny of the mutagenised individuals
Then the DNA samples are pooled to reduce the number of PCR reactions necessary for the detection of a mutation.
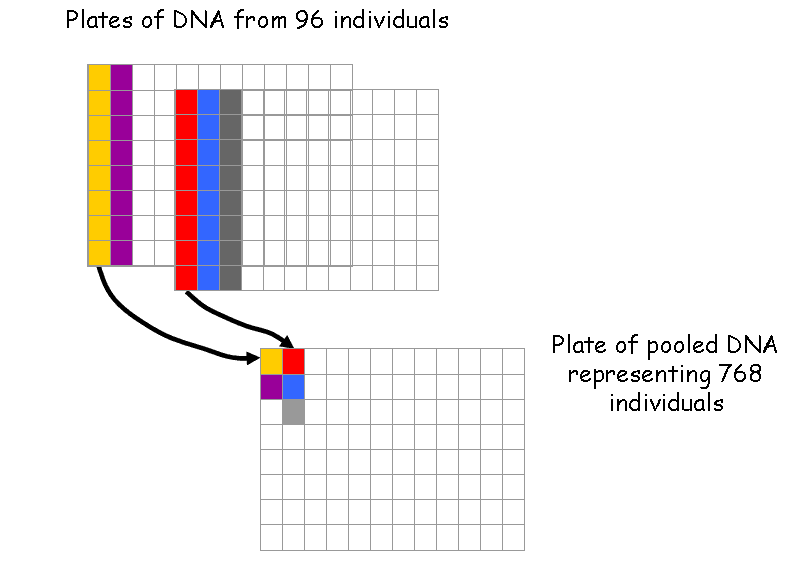
Next the pooled DNA is amplified using fluorescently-tagged, gene-specific primers.
These PCR products are subsequently digested with Celery Juice Extract (CJE) (Till BJ, et al., 2004 Nucleic Acids Res. 32:2632-2641.), which contains an endonuclease that specifically cleaves at the single base pair mismatches that result from heterozygous point mutations, or polymorphisms.
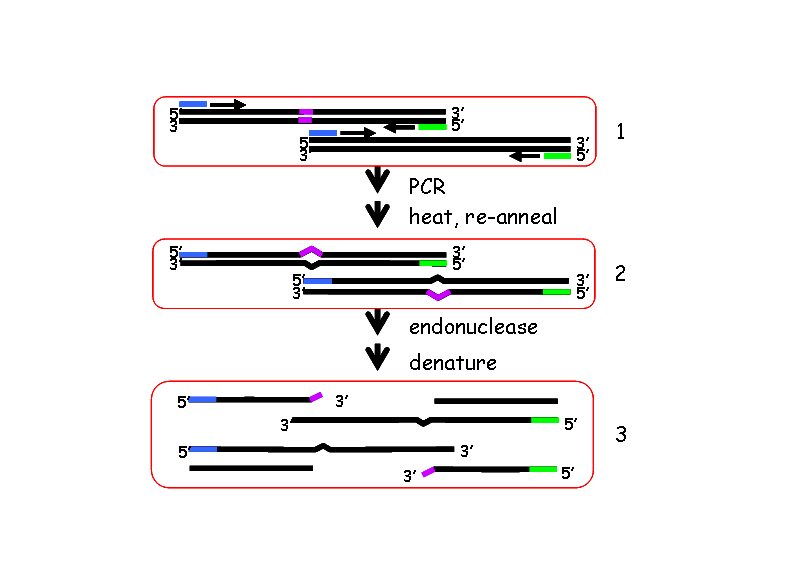
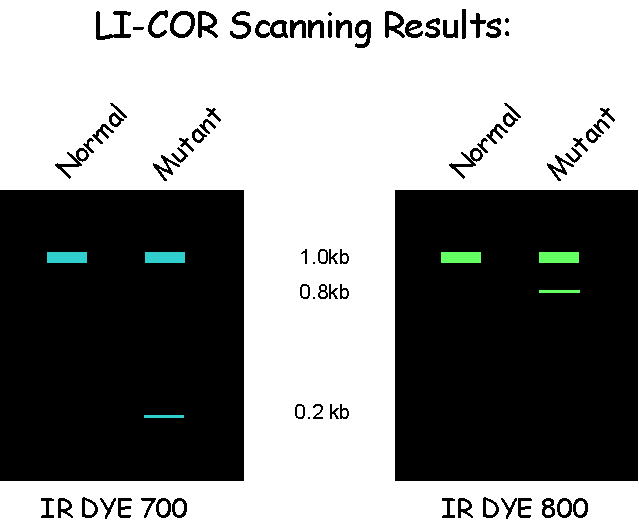
The digested DNA is run on a denaturing, polyacrylamide Li-cor gel in order to identify DNA populations that carry a mutation in the gene being tested. The position of the mutation within the amplicon can be calculated from the size of the complementary bands labelled with the 3′ and 5′ fluorescent tags.
Once a mutation has been identified the procedure is repeated using DNA from individuals that make up the pooled population that exhibited the mutation in the primary screen.





