Microbiologist Michael Baym and colleagues of Harvard Medical School has developed a new experimental setup to watch how bacteria adapt the drug and evolve antibiotic resistance, reported in the Sept. 9 Science. The experimental setup pictures shows step by step how could those minute creatures found in gardens grow up to strong drug fighters.
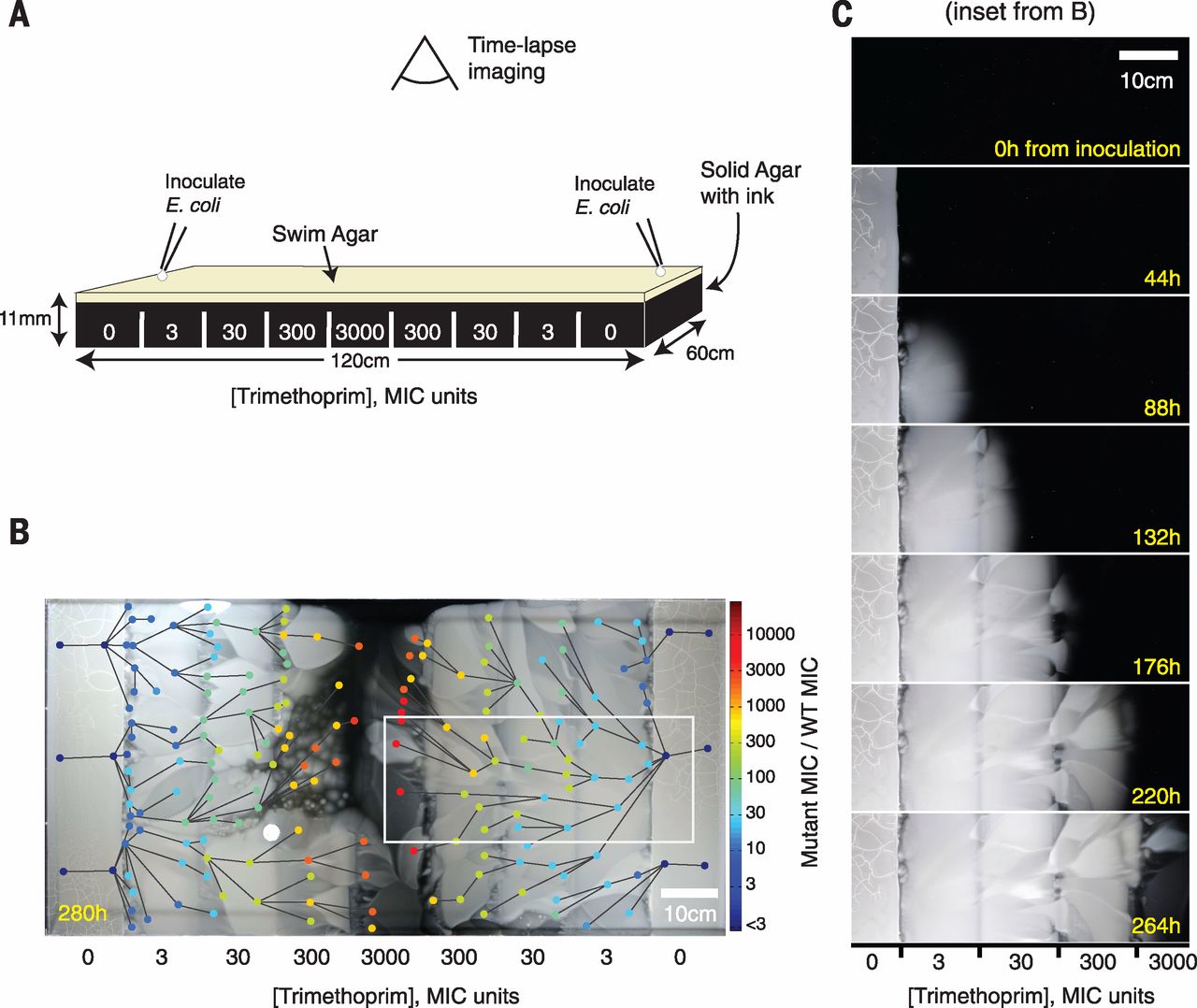
Fig. 1 An experimental device for studying microbial evolution in a spatially structured environment.
(A) Setup of the four-step gradient of trimethoprim (TMP). Antibiotic is added in sections to make an exponential gradient rising inward. (B) The four-step TMP MEGA-plate after 12 days. E. coli appear as white on the black background. The 182 sampled points of clones are indicated by circles, colored by their measured MIC. Lines indicate video-imputed ancestry. (C) Time-lapse images of a section of the MEGA-plate. Repeated mutation and selection can be seen at each step. Images have been aligned and linearly contrast-enhanced but are otherwise unedited.
It is very common for scientists to study bacteria in petri dishes or flasks, a small closed space which includes all experimental materials together, and see how bacteria develop mutant to face and overcome the new challenge of environment.
Although Baym and colleagues did something different as they said, “Inside that flask, in order for a new strain to evolve, the new mutant has to be more fit than everything around it. But in nature, we see a second dynamic: You don’t necessarily need to be more fit than everything around you. You just need to make it into a new environment.”
In order to simulate the natural environment better, they modeled a environment for diversity, an experimental device called the microbial evolution and growth arena (MEGA)–plate was used. By placing different concentrations of trimethoprim or ciprofloxacin (both are widely-used antibotics) in different parts, some of the Escherichia coli bacteria was then observed to have incredible ability to endure a thousand times concentrated antibiotics. However, this sometimes also makes the bacteria spread slowly, which means it may not make them become more competitive in nature selection.
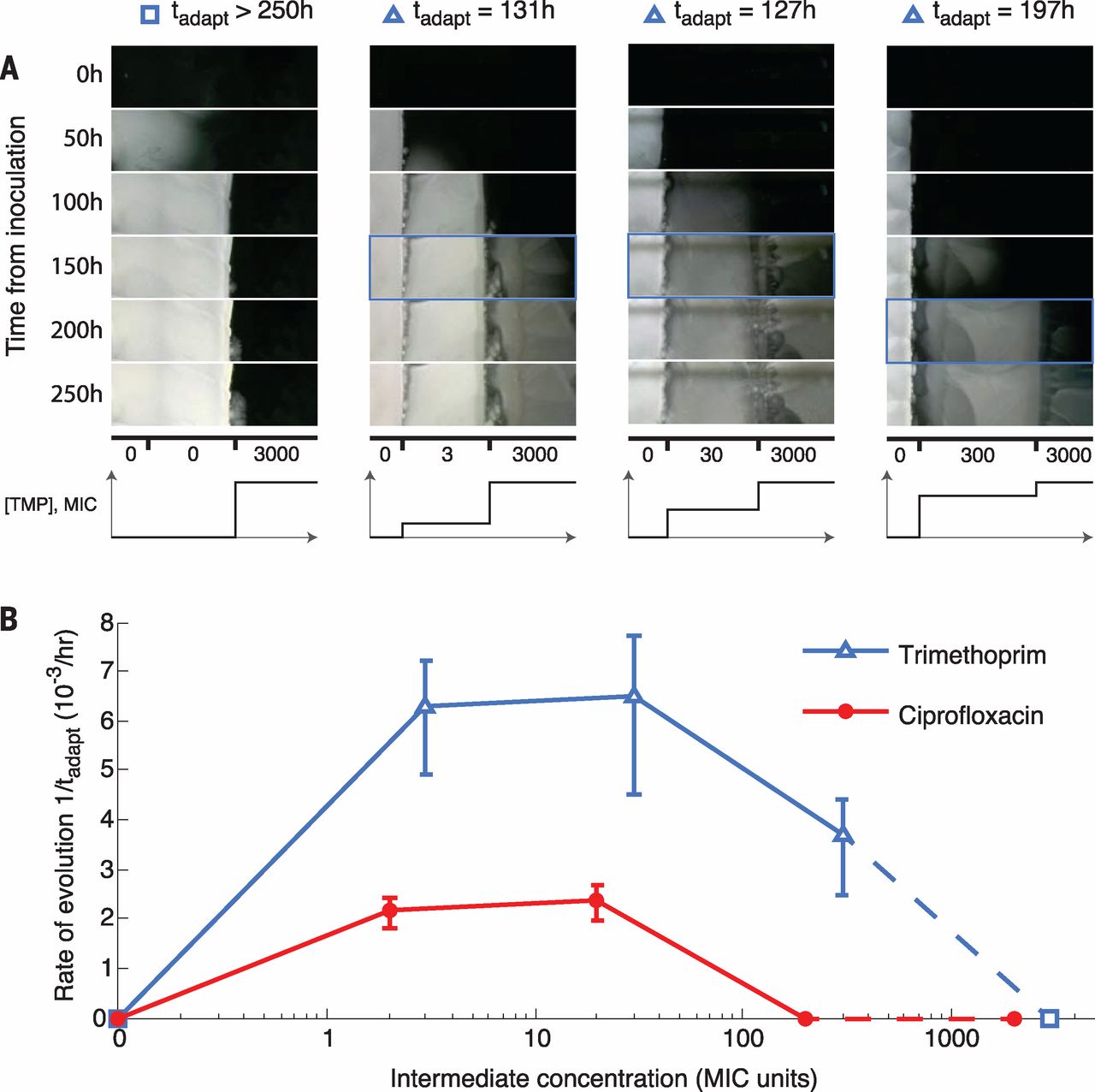
Fig. 2 Initial adaptation to low drug concentrations facilitates later adaptation to high concentrations.
(A) Frames from a section of the TMP intermediate-step MEGA-plate over time (TMP, movie S4; CPR, movie S5). The first frame showing a mutant in the highest band is indicated by a blue box. (B) Rates of adaptation in the intermediate-step experiments across TMP and CPR, showing the necessity of intermediate adaptation for the evolution of high levels of resistance. Error bars show the appearance times of multiple lineages in the highest concentration. Because the intermediate step with no drug puts the highest and lowest concentrations adjacent, it serves as both the highest and lowest intermediate steps (dashed line).
Sam Brown, a microbiologist at the Georgia Institute of Technology in Atlanta who was not in Baym’s group, believes that the bacteria are “climbing this impossible mountain of antibiotics.” Baym and his colleagues thinks this experimental setup could be useful for microbial researches interested in particular environment with special restrictions.
This research suggest that the traditional method is not always the best for new experimental conditions, the new methodology can be developed and applied to fit the real situation. The relative simplicity and ability to visually demonstrate evolution makes the MEGA-plate a useful tool for science education.
Video:
Reference:
M. Baym et al. Spatiotemporal microbial evolution on antibiotic landscapes. Science. Vol. 353, September 9, 2016. doi:10.1126/science.aag0822
