Did you know that not all types of bacteria in this world are harmful to humans? Some bacteria can be used in the production of cheese, and can even be used to manufacture antibiotics for human use. Dr. Jason Hedges and Dr. Katherine Ryan of the University of British Columbia took a look into finding new ways to produce antibiotics from bacteria.
Their recent publication in 2019 showed how they were able to convert an amino acid (a building block of proteins) into a modern day antibiotic.
Antibiotics are used to cure bacterial infections by killing bacteria. For example, an antibiotic called penicillin was used extensively in the 19th century to save thousands of lives.
As Amazing as they Sound, Antibiotics are not a Permanent Solution
The over-usage of antibiotics results in bacteria that can build resistance to them and cause the medication to be ineffective.
The World Health Organization announced: “A serious threat is no longer a prediction for the future, it is happening right now in every region of the world and has the potential to affect anyone” on development of drug resistant bacteria.
Once a type of bacteria develops a resistance to a specific antibiotic, other antibiotics must be used in its place to treat the infection.
Nitroimidazoles are a group of antibiotics which have been used for decades. Due to its low resistance by bacteria, it has been used extensively to treat bacteria that have developed a resistance to certain antibiotics.
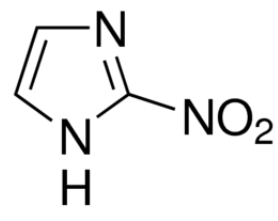
This is the chemical structure of azomycin. Azomycin is a type of antibiotic that falls under the family of antibiotics labelled nitroimidazoles. It is used to treat drug resistant bacteria.
Diving into the Research
The researchers used bioinformatics, a data analysis tool, to find all previous work that has been done on this topic. Bioinformatic searches are performed on databases such as the NCBI.
They found that back in 1953, azomycin (a type of nitroimidazole) was produced by a strain of bacteria that is found in the soil. The researchers adapted this old method by applying modern techniques to it, in an attempt to produce azomycin in a different strain of soil bacteria called Streptomyces cattleya.
The two researchers aimed to find new ways to synthesize azomycin, and proposed that it could be done by using an amino acid called arginine. Once a pathway connecting arginine to azomycin had been developed, their end goal was to synthesize and extract azomycin in Streptomyces cattleya.
Hedges and Ryan were able to develop a multi-step pathway for the conversion of arginine to azomycin through experimentation. However, they were unable to detect any azomycin in the bacteria itself.
They noted that the bacteria of interest was unable to synthesize azomycin, and a separate drug or antibiotic may have produced in its place.
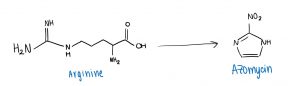
A simplified overview of converting an amino acid (Arginine) into an antibiotic drug (Azomycin).
A Bright Future
The significance of this research transcends the synthesis of azomycin. Although they failed to detect azomycin in Streptomyces cattleya, their work provides a stepping stone for further research to be conducted.
This study provides insight on new biosynthetic pathways which is important to those currently in the field of life sciences and pharmacology.
Furthermore, this study expands people’s knowledge of bacteria engineering and how antibiotics are created.
In the near future, researchers could test their synthetic pathway on other bacteria such as E. coli to determine if azomycin could be produced. This will allow for the production of more antibiotics, or even the discovery of new antibiotics.
Reference:
Hedges, J. B.; Ryan, K. S. In Vitro Reconstitution of the Biosynthetic Pathway to the Nitroimidazole Antibiotic Azomycin. Angewandte Chemie International Edition 2019, 58 (34), 11647–11651.
– Adrian Emata, Vicky Gu, Jackson Kuan, Yicheng Zhu
