Although eating too much sugar can lead to health complications, a normal intake of sugar has its benefits. This is because sugars take part not only in many cellular activities as an energy source but also in cell-cell communication as a communicator. In 2019, a research team led by Dr. Stephen Withers at the University of British Columbia made different sugars by using enzymes (Pac-Man-like molecules) in bacteria. This finding enables the building of many sugars that are commonly hard to find in nature.
What are sugars?
All sugars are made of hexagonal building blocks as shown in red in Figure 1. Two common building blocks are glucose and fructose. To make different sugars with only two common building blocks, we can vary the number and arrangement of the building blocks. For example, cellulose has three long threads that are arranged almost parallel to each other. In contrast, starch has one long thread that adopts a helical structure.
Now we know that different sugars can be made by varying the kinds of building blocks, the numbers of building blocks and the arrangements of building blocks. What we haven’t talked about is how the building blocks are linked. There are two issues with linking the building blocks. First, building blocks are much smaller than our cells. Getting two building blocks together is as hard as fishing a needle from the Pacific Ocean. Besides, getting two building blocks together takes time because they prefer to be alone. How can our mother nature solve these problems to keep us alive?
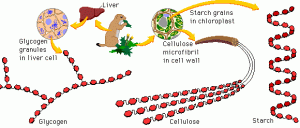
Figure 1. Sugars are built differently. Some sugars are longer and more complex than others. Source: riasparklebiochemistry
Here comes the rescue…Pac-Man in the cells!
The illustrations of enzymes are like Pac-Man. However, different from Pac-Man that eats any “food” in different shapes, enzymes recognize different substrates and those substrates only! Because of this substrate-enzyme specificity, linking two building blocks together is much easier. As shown in Figure 2, the enzyme will attract two specific building blocks, making them closer to each other and eventually join.
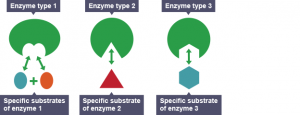
Figure 2. Different enzymes recognize different substrates. Source: Wikipedia
Besides, enzymes can speed up the “hugging” process between the substrates (building blocks), making the formation of a long sugar favorable.
Figure 3. Enzymes speed up reactions. Source: pinimg.com
Manipulation of enzymes to build different sugars for cell-cell communication
Knowing that enzymes have many advantages, Dr. Wither and his team looked for enzymes that speed up the linkage of sugar building blocks in E. Coli, bacteria that live inside our digestive tracts. They chose eight enzymes out of the 175 sugar-specific enzymes in E. Coli. These eight enzymes were the most specific to the sugar building blocks they were interested in. However, after further investigation, the researchers found that the enzymes helped speed up the breakage of the linkage(s) between sugar building blocks instead of the formation of the linkage(s).
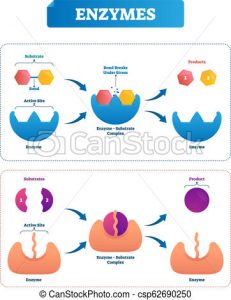
Figure 4. Some enzymes speed up the breakage of linkages while others speed up the formation of linkages.
Source: Canstock.com
To solve this problem, the researchers reverse-engineered these enzymes from speeding up breakage to speeding up linkage by changing parts of the enzymes. Now different sugars can be made! As mentioned above, cell-cell communication relies on sugars. This is because two cells adhere via the interaction between extracellular sugars and specific cell-surface receptors. Once these cells adhered to each other, they can talk to each other about their internal cellular condition/environment and trigger a corresponding response. This has great implications in triggering an immune response to a viral attack. By engineering more sugars on the surface of the cells, cells in our immune system can more quickly talk to each other and fight off infection.
Journal Source:
Armstrong, Z.; Liu, F.; Chen, H.-M.; Hallam, S. J.; Withers, S. G. Systematic Screening of Synthetic Gene-Encoded Enzymes for Synthesis of Modified Glycosides. ACS Catalysis 2019, 9 (4), 3219–3227.
-Pricia Ouyang
