Can it really be that simple? Can the answer really be in the ground beneath our feet?

Source: Soil Science Society of America
Discovery of a gene cluster commonly found in soil-dwelling bacteria may be the key to treating anaerobic bacterial infections such as appendicitis and pneumonia. Researchers Jason B. Hedges and Prof. Dr. Katherine S. Ryan from the University of British Columbia have isolated the antibiotic compound azomycin from a biosynthetic gene cluster found in the bacterium Streptomyces cattleya.
With this information they believe it can lead to engineering bacteria to produce a new line of antibiotics.
What is a gene cluster?
The term gene cluster is moreso semantics to describe a group of genes that share a common phenomenon.
Originally, azomycin was isolated from a similar bacterium Streptomyces eurocidicus back in 1953 and became the blueprint to synthetic nitroimidazoles.
“Nitroimidazoles are one of the most effective ways to treat anaerobic bacterial infections”,[1] Hedges writes in his 2019 study. The most commonly used nitroimidazole, metronidazole, is an antibiotic used to treat pelvic inflammatory disease, endocarditis, and bacterial vaginosis.[2] It is also on the World Health Organization’s List of Essential Medicines, the safest and most effective medicines needed in a health system.[3]

Figure 1: Molecular Structure of Nitroimidazole. Source: Sigma Aldrich
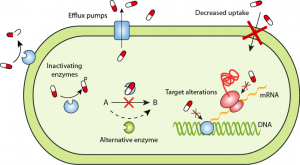
Despite its use in antibiotics for over 60 years, the incidence of nitroimidazole resistance in anaerobes remains low, making it an essential component of the antibiotic arsenal. [1,4]
Since the isolation of azomycin back in 1953, synthesis of nitroimidazoles were limited to synthetic routes, most commonly involving reactions of an imidazole with nitric acid and sulfuric acid.
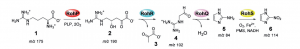
Using bioinformatics, however, in 2019 Hedges and Ryan were able identify a biosynthetic gene cluster in the same bacterium that makes penicillin,[5] Streptomyces cattleya. They were able to find that this gene cluster containing azomycin is widely distributed among soil-dwelling actinobacteria and proteobacteria.
Because of this they theorize that azomycin and other nitroimidazoles may be important factors in ecology.
What are bioinformatics?
Bioinformatics is the science of collecting and analyzing complex biological data such as genetic codes. As an interdisciplinary field of science, bioinformatics combines biology, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data
In addition to the isolation of azomycin in a gene cluster, Hedges and Ryan were able to perform in vitro analysis in order to understand the enzymatic steps that take the primary protein L-arginine to become azomycin.

Source: Washington Post
Their work opens the door to biocatalytic methods to synthesize azomycin and other nitroimidazoles. They believe this discovery can “lead to the possibility of engineering bacteria to produce nitroaromatic compounds”.[1]
In other words, this may lead to stronger antibiotics immune to drug resistance.
References
Hedges, J. B.; Ryan, K. S. In Vitro Reconstitution of the Biosynthetic Pathway to the Nitroimidazole Antibiotic Azomycin. Angewandte Chemie International Edition 2019, 58 (34), 11647–11651.
The American Society of Health-System Pharmacists. Archived from the original on 6 September 2015. Retrieved 31 July 2015.
World Health Organization model list of essential medicines: 21st list 2019. Geneva: World Health Organization
David I. Edwards, Nitroimidazole drugs-action and resistance mechanisms I. Mechanism of action, Journal of Antimicrobial Chemotherapy, Volume 31, Issue 1, January 1993, Pages 9–20, https://doi.org/10.1093/jac/31.1.9
Kahan, JS; Kahan, FM; Goegelman, R; Currie, SA; Jackson, M; Stapley, EO; Miller, TW; Miller, AK; Hendlin, D; Mochales, S; Hernandez, S; Woodruff, HB; Birnbaum, J (Jan 1979). “Thienamycin, a new beta-lactam antibiotic. I. Discovery, taxonomy, isolation and physical properties”. The Journal of Antibiotics. 32 (1): 1–12
-Adrian Emata