[There was misformatting in while transferring the document, so the numbers are mixed ]
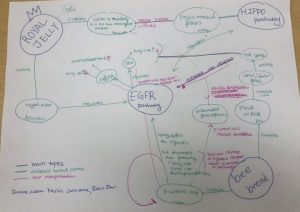
This is one of my top 4 assignments because the lonfat paper was the hardest paper in this class in that they had multiple techniques and had a complex way of studying gene interaction. I initially had no idea what they were doing with the inversions and what they were trying to say in the paper. After having discussions in class, it became clearer that the inversion would change the position of the genes, and therefore would allow us to study whether or not the position of enhancer affects gene expression. From this paper, I learned how to read the bisulfite sequencing paper, and how to think about different data (staining, bisulfite sequencing, experimental system, and chip) can come to one interpretation. It was interesting to learn that enhancers can be position dependent in some cases.
In-class small assignment: Lonfat et al. paper
Assigned paper: Lonfat, N., Montavon, T., Jebb, D., Tschopp, P., Nguyen Huynh, T.H., Zakany, J., Duboule, D. (2013). Transgene- and locus-dependent imprinting reveals allele-specific chromosome conformations.
Supporting PDF: http://www.pnas.org/content/pnas/suppl/2013/06/30/1310704110.DCSupplemental/pnas.201310704SI.pdf
Names of your group members:
Shannah Fisher, Joanne Lim, Erin Yang
Question 1
It is quite common for research papers to have Figure 1 be the “most important” figure in the article. Consider Figure 1:
- What transgenic lines did the author use? Please briefly describe them (do these lines look somewhat familiar)?
The transgenic lines used were Hox9lacZ transgene inserted into the HoxD gene in both the forward direction and with an inversion in the region starting from ex26 to the end of Hoxd9lacZ insertion (contains ex26, Pdk1, digit enhancer sequences and the Hoxd9lacZ insertion).
(HoxD9lacZ transgene was inserted upstream of the HoxD gene. One transgenic line had HoxD9lacZ closely upstream of HoxD, and the other transgenic line had an inversion at Itga6loxP and rel5loxP such that the transgene is located further away from Hox D.
The transgenic lines shown in the Lonfat et al. paper is similar to that shown in class in that both are designed upstream of HoxD. However, the transgenic line from class includes Evx2 in the inversion.)
- What do the data in Panel C show?
In the normal HoxDrel5 embryo’s, there is no difference in expression of lacZ transgene between maternal and paternal inherited transgenes. However, in the inverted HoxDInv(rel5-Itga6) transgene, the embryo’s with paternal inherited transgenes showed higher expression of lacZ insert (compared to HoxDrel5 embryos) and maternally inherited trangenes showed no expression of lacZ.
- What is striking/unexpected about the data in Panel C?
When the Hoxd9lacZ inversion is inherited through the paternal line, fetuses show very strong staining from the transgene beyond its typical regions whereas the identical transgene inherited through the maternal almost entirely lacks staining. This suggests a very efficient regulatory mechanism that exists as separate inheritable paternal and maternal patterning.
- What direct conclusions do you make from the data?
Inversion of the region is sufficient to increase the expression of lacZ in the embryo with paternal inherited transgene as well as decrease expression of lacZ in maternal inherited transgenic embryos.
Question 2
How did the authors show that the peculiar effect observed is specific and position/site-specific?
- The authors first inserted the lacZ transgene in the HoxDrel5 (rel5) site and observed no bias in expression based on parental inheritance.
- After inverting the region using the loxP sites in rel5 and exon 5 of Itga6, they observed that the lacZ transgene was now biased in expression depending on which parent the transgene was inherited from.
- The translocation results suggest regulatory effects influenced by parental inheritance that differ between Itga6 site and the rel5 site.
- The authors showed that the result is transgene specific because the same effect was not observed when tested with SB lacZ.
Do you agree with their data interpretation and with their conclusion?
- Yes, in the case of the lacZ transgene it seems that the effects are site specific. But the in vitro relevance of this data needs further development as a Hoxd11 inversion without the lacZ transgene didn’t produce the same bias in expression.
Question 3
Consider Figure 2 (please don’t hesitate to ask for clarification if you and your group have questions about it!):
- Briefly explain how to “read” the diagrams shown (i.e. what do the rows of circles represent, what do the white vs. black circles represent).
- The white dots represent unmethylated regions, and the black dots represent methylated regions. Each dots represent DNA regions.
- What do the data in Figure 2 show?
- Embryos that paternally inherited transgenic line HoxDInv(rel5-Itga6) showed very low levels of methylation of DNA from E12.5 presumptive digits while those that maternally inherited HoxDInv(rel5-Itga6) showed extremly high methylation of DNA.
- The escapers showed varying methylation patterns.
- DNA from oocyte with the inverted allele is highly methylated while from the sperm isn’t. This suggests that the methylation patterns are carried through the oocyte and to maternally inherited offspring.
- Methylation patterns are not different in embryos that inherited rel5.
- Why aren’t there a “paternal/+” and a “maternal/+” groups for sperm and oocytes?
Sperm and oocytes are haploid gametes, each containing one allele. There is no paternal or maternal allele in the gametes.
- What are “escaper” embryos, and how were they identified prior to bisulphite sequencing?
Escaper embryos are maternal transgenic embryos that expressed variable levels of lacZ unlike other maternal inherited embryos that had no expression. These embryos expressed it despite the allele being silenced through maternal imprinting, thus “escaping” silencing. Expression of lacZ was studied through in situ hybridization and lacZ staining.
- What can we directly conclude from the data?
High levels of DNA methylation is inherited through the oocyte germline. Inversion in the transgenic line is sufficient to result in extremely different methylation patterns with paternal/sperm DNA having low DNA methylation while maternal/oocyte having high DNA methylation.
Question 4
Figure 3 depicts the results of a series of 4C experiments. Try to “read” the figure and see if you can identify the information described in the text.
- What did the authors do, and what are the results?
They use the lacZ sequence as bait for 4C experiments in normal HoxD-rel15 and inverted HoxDInv(rel-15-Itga6) for both paternal and maternal inherited transgenes.
Before inversion, there is no difference in interactions between lacZ and enhancer regions between the paternal and maternal inherited trangenes. However, after inversion, the paternal and maternal inherited transgenes, show a difference in interactions between enhancers and lacZ.The authors discuss that only the paternal allele interacts with the transgene and the digits enhancers, suggesting further effects on regulatory interactions based on the parental imprint.
- What can be directly concluded from the data?
The inversion of the rel5-Itga6 region is sufficient to change the interactions between lacZ with maternal and paternal inherited transgenes.
Question 5
How does each figure support the statement in the title of the article? Which one supports it most?
Transgene dependent vs locus dependent imprinting → allele specific chromosome conformation
Figure 1 & 2: shows that maternal imprinting is dependent on the inversion, which brings the Hoxd9lacZ closer to Itga6. They showed transgene dependent imprinting by doing the same experiment with different transgenes.
Figure 3: reveals allele-specific chromosomal conformation by showing 3D conformation through 4C experiment, so this is figure supports the title the most.