Enhancers and Gene Regulation
When it comes to transcription there is still a lot we don’t fully understand about how it works. Although we don’t fully understand transcription regulation, there is a lot of exciting research happening in order to shed light on the process. Transcription is a sporadic process, bursts of transcription results in mRNA production followed. We talked in class about the role of enhancers in gene regulation. A study done by Fukaya, Lim and Levine demonstrated that enhancers affect gene regulation by controlling the number of bursts that occur and therefore the amount of mRNA that is produced (2016). Strong enhancers increase the burst frequency such that transcription is continuous and there is little time between bursts. Weak enhancers have the opposite effect and cause transcription to occur is sporadically. Before discussing gene regulation in class, I had never heard about transcription bursts and I was pretty intrigued and did a little more reading on the subject. In my head, whenever I thought about transcription regulation I pictured an enhancer element “attracting” a polymerase enzyme to the promoter in order to express the gene. I thought a strong enhancer was able to attract more polymerase enzymes to the promoter than a weak enhancer in a given amount of time. This new model of gene regulation was really interesting to me because I had never thought about transcription as occurring in bursts, I thought it was a continuous process. The following diagram, illustrates how enhancers affect gene regulation.
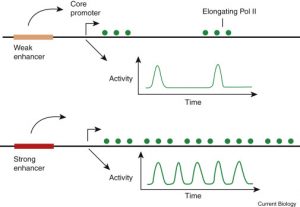
(Muerdter & Stark, 2016)
Work Cited:
Fukaya, T., Lim, B., & Levine, M. (2016). Enhancer control of transcriptional bursting. Cell, 166(2), 358-368.
Muerdter, F., & Stark, A. (2016). Gene Regulation: Activation through Space. Current Biology, 26(19), R895-R898.
Imprinted Gene Expression
I think it’s really interesting how imprinted gene expression is dependent on the parent-of-origin and because I think imprinted genes are super cool, I decided to study them for my final project! I did quite a bit of research on the Airn/Igf2r imprinted gene cluster. Igf2r gene expression is repressed by Airn in a transcription-dependent manner, this means that the Airn transcript does not interact with Igf2r and simply the act of transcription represses Igf2r expression. This makes sense because the Airn gene is located on the antisense to Igf2r. Airn also regulates the expression of over imprinted genes in a transcription-independent manner (Slc22a2 and Slc22a3). One of the questions I had based on my readings was: what dictates whether an imprinted gene is regulated in a transcription-dependent or a transcription-independent manner? For example, Mas1 is located on the antisense strand of Airn (Mas1 is located downstream from Igf2r), if transcription-dependent regulation occurs as a result of genes overlapping then why isn’t Mas1 an imprinted gene?
X-Chromosome Inactivation
Although in previous classes we’ve talked a little about X-chromosome inactivation, in this class we really dove into it. One of the really interesting discussions we had in class was regarding which organism would be a good model to study X-chromosome inactivation as different species have slightly different X-chromosome inactivation mechanisms. Specifically, we compared X-inactivation in marsupials, mice and humans. Marsupials inactivate their paternal X-chromosome in all tissues, mice and humans also inactivate their paternal X-chromosome early in development but later on, X-inactivation is random and either the maternal or the paternal X-chromosome is silenced. If we’re interested in studying X-chromosome inactivation in humans, then studying human embryos would be the best model system to use. However, because we are not yet manipulating human embryos, I think that working with a murine system is the best model system to use to study X-inactivation. Mice embryos undergo imprinted X-chromosome inactivation as well as random X-inactivation like human embryos and it is likely that the mechanism for X-inactivation was past down from a shared ancestor. Thus the mechanism of X-chromosome inactivation is likely conserved and studying a murine model can potential shed light on the human system. Additionally, mice are used as model systems to study other processes and we know that humans and mice share similar genes.
CRISPR/Cas9 Gene Editing System
Since it was first developed to edit the genome, CRISPR/Cas9 has been a highly controversial topic. CRISPR/Cas9 is a revolutionary system that allows researchers to edit the genome of organisms with a high level of specificity. Using CRISPR, researchers can alter genes by making precise nucleotide base changes and can even insert/delete DNA sequences. This genome editing system is not perfect, and one of the biggest issues with CRISPR/Cas9 is off target effects. CRISPR/Cas9 works by targeting a Cas9 enzyme to a specific sequence to create a double stranded break in the DNA. If Cas9 is targeted to the wrong sequence, it will produce double stranded breaks at unwanted sites throughout the genome. I did my Technique Infographic on the TALENs system which is another gene editing system. Through this project I read about the CRISPR/Cas9 system and I found some interesting papers on the work scientists are doing to engineer a CRISPR/Cas9 system that has little to no off target effects. One of the engineered systems that I thought was particularly interesting, is an engineered CRISPR system that mimics the TALENs system. Scientists have engineered Cas9 enzymes so that they can no longer make double stranded breaks. These Cas9 enzymes can still be targeted to a sequence but they cannot edit the genome. By hybridizing these engineered Cas9 enzymes to an endonuclease such as Fokl that is only active in a dimer state, researchers are able to introduce a high level of specificity into the system. For this Cas9-Fokl system to be able to edit the genome, two molecules of Cas9-Fokl have to bind to the target DNA sequence in order for Fokl to become active. Two molecules of Cas9-Fokl are target to different sequences that are near each other. Once the Cas9 molecules bind to their sequences, the Fokl endonucleases can dimerize and make a double stranded break. Because two Cas9 enzymes have to bind different sequences that are near each other for a double stranded break to occur, this adds a high level of specificity to the system. I think it is amazing the kind of work researchers are doing. If, in the future we are able to eliminate the off target effects of the CRISPR/Cas9 system I think that there is a potential for this system to be used for germ line gene editing. CRISPR/Cas9 could have a huge impact on reducing the risk of serious diseases within the population.
RNA/DNA FISH Protocol:
In class we often analyzed results collected from RNA/DNA FISH experiments. For my final project, I designed an RNA/DNA FISH experiment in order to study the localization pattern of Airn RNA relative to the Tcp-1 and Sod2 loci. I found the following video by jOVE on the RNA/DNA FISH protocol to be very helpful in understanding how RNA/DNA FISH experiments work, and the kinds of data you can collect using this kind of an experiment.