i) I analyzed the Spannhoff et al paper with my partner, Melissa Liu. The paper investigated the ability of 10HDA, a lipid found in Royal Jelly, to epigenetically modify genes. They found that it had histone deacetylase inhibitor activity, and may be responsible for some of the epigenetic effects of Royal Jelly. A copy of our analysis of the article can be found here.
Spannhoff, A., Kim, Y.K., Raynal, N.J., Gharibyan, V., Su, M.B., Zhou, Y.Y., Li, J., Castellano, S., Sbardella, G., Issa, J.P., Bedford, M.T. (2011). Histone deacetylase inhibitor activity in royal jelly might facilitate caste switching in bees. EMBO Reports, 12(3), 238-43. doi:10.1038/embor2011.9
ii) Here is a copy of our group’s model:
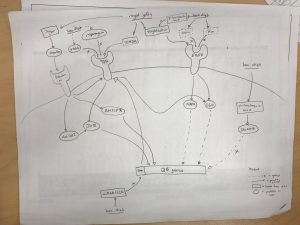
Group members: Evan Gibbard, Hayden Wood, Melissa Liu, Miguel Oreta, Oline Pade Jensen
iii) A document with the class’ models for Queen bee development can be found here.
iv) Summary of the model: The class generally agreed that components of Royal Jelly (e.g. royalactin) upregulated the EGFR pathway, while components of Bee bread (e.g. vein, km) downregulated the EGFR pathway. The EGFR pathway induced Queen bee phenotype through downstream pathways (e.g. mapk, S6K) that induced Queen bee phenotype.
Contradicting evidence was found in the paper “Royalactin is not a royal making of a queen” which found that royalactin alone was not enough to induce Queen bee phenotype, and had no measurable effect on influencing phenotype.
Spannhoff et al. also found that the large fraction of Royal Jelly (components larger than 3kDa) could not induce epigenetic modifications in their system, and did not think that proteins could be responsible for the epigenetic effects of Royal Jelly.
One way of testing this would be through the creation of a synthetic royal jelly with all necessary components for Queen bee phenotype development. Once the synthetic RJ is shown to be sufficient to induce QB phenotype, we could selectively remove components of RJ or a combination of different components to see which are necessary for the development of QB phenotype.
Another way of testing this would be by identifying which factors influence the larvae towards the QB or worker bee phenotype. For example, 10HDA and royalactin would be group together as QB factors, and the plant miRNAs and vein + km would be grouped together as worker bee factors. This would work well with the synthetic Royal Jelly experiment to determine which factors induce caste differentiation.
v) Reference list:
Buttstedt, A., Ihling, C. H., Pietzsch, M., & Moritz, R. F. (2016). Royalactin is not a royal making of a queen. Nature,537(7621). doi:10.1038/nature19349
Ashby, R., Forêt, S., Searle, I., & Maleszka, R. (2016). MicroRNAs in Honey Bee Caste Determination. Scientific Reports, 6, 18794. http://doi.org/10.1038/srep18794
Kamakura, M. (2011). Royalactin induces queen differentiation in honeybees. Nature, 473(7348), 478-483. doi:10.1038/nature10093
Mao, W., Schuler, M. A., & Berenbaum, M. R. (2015). A dietary phytochemical alters caste-associated gene expression in honey bees. Science Advances, 1(7), e1500795. http://doi.org/10.1126/sciadv.1500795
Spannhoff, A., Kim, Y.K., Raynal, N.J., Gharibyan, V., Su, M.B., Zhou, Y.Y., Li, J., Castellano, S., Sbardella, G., Issa, J.P., Bedford, M.T. (2011). Histone deacetylase inhibitor activity in royal jelly might facilitate caste switching in bees. EMBO Reports, 12(3), 238-43. doi:10.1038/embor2011.9
Zhu, K., Liu, M., Fu, Z., Zhou, Z.,. Kong. Y., H., Lin, Z., Luo, J., Zheng, H., Want, P., Zhang, J., Zen, K., Chen, J., Fuliang, H., Zhang, C., Ren, J., Chen, X. (2017.) Plant microRNAs in larval food regulate honeybee caste development. PLOS Genetics 13(8), e1006946.