Top 1/4: In-Class Assignment
In-Class Assignment: Oct 21st, 2015
I chose this assignment specifically for question 4b. My group had a really great discussion about this topic. It reminded me of a course I took the other summer semester, IHHS 400 which was about working in multidisciplinary health care teams and about interacting with patients of the family. I think in any research it is important to always consider the big picture and in health-related research where human patients are being used it is important to be aware of the impact this plays in their life. I really enjoyed this article as well because I was learning about Hox genes in BIOL 331 around this same time so it was interesting to see research being done on principals I was learning in other classes.
Top 2/4: In-class Assignment
In-Class Assignment : Sept 21st, 2015
Even though this assignment was completed before my the in-class assignment of October 21st that I also chose as in my top 4 assignments, I chose to include this assignment based on having included the one from October 21st. When searching through my work for a final “top 4” assignment, I found this as one of the first things we did in BIOL 463 this term. I was looking over the answers my group gave at the time and though I think we did a great job, I would answer these questions so differently now having completed the term. I chose this assignment because it shows the progression of my learning. At the time along with my group, I had a lot of difficulty navigating a scientific paper, finding the terminology as one of the most difficult aspects. I find myself thinking – go look at the sources of the paper or go review the graphs! Now the thing I would find more difficult is would be perhaps whether or not a graph sufficiently showed the claims in the title rather than just data go
News and Views Presentation
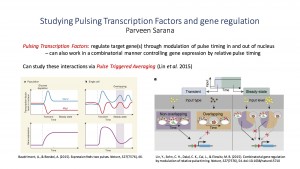
Transcription factors are proteins that bind to specific DNA sequences such that they control the rate of transcription from DNA to RNA. Many transcription factors act continuously, but there are some transcription factors that work in “pulses” of moving quickly in and out of the nucleus. By working in pulses, these pulsing transcription factors offer more information like their frequency of pulsing and have a more complex regulation process on the genes they regulate (Lin et al., 2015). Studying these pulsing transcription factors is not simple because the way these pulsing transcription factors act is very different from cell to cell. Lin et al. adapted a neuroscience technique known as spike-triggered averaging to develop a method known as pulse-triggered averaging and studied the Msn2 and Mig1 transcription proteins Saccharomyces cerevisiae.
Pulse triggered averaging is essentially a way to standardly study pulses of transcription factors around a set peak. In this case, pulses are measured as averages based on the interactions of Msn2 and Mig1 over a set time period around peaks in the nuclease of the Msn2 levels. Msn2 is known to act as an activator when it inside the nucleus and Mig1 is known to repress transcription of the same genes – both these proteins are pulsing transcription factors (Lin et al., 2015).
By creating strains of this yeast where both Msn2 and Mig1 were tagged by fluorescent proteins, they tracked the proteins intracellular location over time. They studied a single cell at a time instead of entire populations by subjecting the cells to various external stimuli such as depleting and increasing levels of glucose in the cell growth medium all while monitoring the location of the Msn2 and Mig1. Lin et al. found that there was a change in the way the pulsing of Msn2 and Mig1 in response to external stimuli (2015). Decrease in glucose resulted in large transient decrease in the level of Mig1 in the nuclease but an increase of Msn2 in the nuclease. This causes what is known as an “overshoot” allowing the cells to adapt to change quickly by increasing gene expression. After this transient/overshoot stage/event, the transcription factors and gene expression remains steady population-wide. When looking at specific cells, however, Lin et al. found that the behavior was not the same as what was seen population-wide. Only in the transient phase cells display synchronized and non-overlapping pulse of transcription factors Msn2 and Mig1. After this, levels of each of the factors pulses in a random order. When Msn2 and Mig1 overlap, gene expression decreases but when the Msn2 pulse does not overlap with Mig1 the gene expression increases.
By looking at the pulsing transcription levels in a specific cell versus population-wide, it is seen that the pulsing behavior is more complex and interactive between various transcription factors than once thought. This new technique of pulse triggered averaging will open up for new research to be done to look into signaling pathways to a level of detail that was not possible to study before.
Lin, Y., Sohn, C. H., Dalal, C. K., Cai, L., & Elowitz, M. B. (2015). Combinatorial gene regulation by modulation of relative pulse timing. Nature, 527(7576), 54. doi:10.1038/nature15710
Question: Given what you know about how pulsing transcription factors function in relation to external stimuli, what do you predict about the concentration of target gene(s) and transcription factors in the nucleus over time in the following situation:
A single cell of the model organism, Saccharomyces cerevisiae, is studied looking at the pulsing transcription factors Msn2 (a known activator) and Mig1 (a known repressor). The cell is exposed to decreasing levels of glucose and you are looking at specifically the “steady state” time period. Which of the following would you predict to be possibly occurring in the single cell in question at this steady state phase? Select all that apply.
- Increased levels of Msn2 in the cell so there is an overall increase in gene expression level of the target gene.
- Msn2 and Mig1 pulses overlap in the nucleus resulting in decreased transcription level of target gene.
- Msn2 pulse does not overlap with the Mig1 pulse resulting in increased transcription level of the target gene.
- Msn2 and Mig pulses overlap in the nucleus resulting in increased transcription level of target gene.
Answer: Both B and C are possibilities that could be seen in a given cell because the behavior of pulsing transcription factors in individual cells is dynamic and somewhat random so though in the overall population we may see the trend decreased in A, both B and C could be taking place in individual cells. D is a distractor as it lays out the same scenario as B, but with the incorrect result.
See the following section for my reflection on this assignment:
Top 3/4: Learning Journal #3
https://blogs.ubc.ca/parveensarana/learning-journals/
The learning journals were some of the assignments that I found both the most enjoyable and helpful to my learning so I wanted to include a sample of one in my Top 4 Assignments. I think Learning Journal 3 is one of the best ones I did this term. Something that stands out to me about this assignment is the “Evidence/how you would test someone on this” section. I think that developing a question is one of the best ways to see if you really know the material because you have to not only think of the correct answer but possible confusions that could arise to use as distractor points in the question. Something I would change here is that the type of knowledge for my understanding of the 3C technique presented is more conceptual than factual as the question I proposed is more “skill-knowing” rather than just fact based. In my opinion you need to know the factual knowledge before conceptual knowledge can be applied. This learning journal was especially helpful when I was working on my N&V assignment. When creating the N&V assignment question I had to think – what am I trying to test? How can I word a question so that it is testing a higher level of understanding?
Top 4/4: If I were a Developmental Biologist
I chose to include the following as part of my Top 4 Assignments because this was really the starting point of so much that I would learn in this class. Looking back, I’m actually impressed with the level of questions I was able to come up with, specifically question #2. This question actually led me down the path to my project question in a way because I ended up looking at the effect of maternal genotype of offspring so this was a very helpful assignment to get the creative juices going! I’d never really been asked this type of question in other courses before so I chose to include this as it got me interested in this course right off the bat.
1) At what point of development do mutations arise or is the regularly observed developmental process altered in cases of sexual ambiguity?
2) As the placenta acts as an essential link between mother and the developing fetus, could a wider array of antibodies be introduced to the fetus via the maternal circulatory system (and then placenta to fetus) in order to increase the types of antibodies in the fetus immune system when born?
3) Question 2 – This question appeals to me as it sort of bridges various concepts I have learned from developmental biology to microbiology. Ever since I was introduced to the idea of fetal development, I was interested in the role and impact of the mother’s health in that developmental process. We know that many health conditions are caused directly by choices of the mother such as fetal alcohol spectrum disorders, but there seems to be less talk about the positive impact/advantages a healthy mother could have on/give the developing fetus. Later, when I was introduced to the idea of passive immunity and antibodies, I began to question whether or not we could manipulate this naturally occurring process to increase the “power” of the fetus immune system such that once born the immune system would have acquired a wider array of resistance than could have been offered by the mother’s natural antibody library.
If this question were to be answered, it would allow for a new area of study to be opened up. More questions such as to what degree we could manipulate the immune system and what else could be introduced via the placenta interface could be explored. If the question is answered such that this introduction of new antibodies via the placenta is possible, we could work to introduce immunity early on to diseases which would allow for a healthier population overall. This may be seen as a miracle procedure if through further research we could eradicate diseases that affect newborns via introduction of antibodies into their system.This type of manipulation would possibly split the community at large as some may oppose to manipulation of fetal development regardless of positive impact as it may be seen as against nature.