Research
Adaptation and Life History Evolution
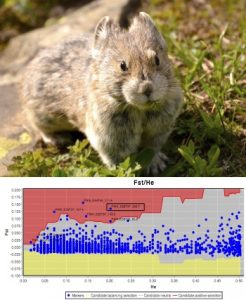 Biologists have long appreciated the importance of preserving the ability of organisms to adapt to changing environments, yet have been limited in their capacity to quantify and use adaptive genetic variation for prioritizing populations for management. Until recently, our ability to directly investigate adaptive genetic variation has been limited to a handful of model organisms studied under artificial conditions. Population genomics combines genomic technologies with population genetics theory to provide a framework for detecting genome-wide associations between segregating variation and fitness-related traits in natural populations. This approach seeks to identify loci that exhibit signatures of selection in comparative genomic scans of populations exhibiting varying phenotypes and/or relative to environmental variation.
Biologists have long appreciated the importance of preserving the ability of organisms to adapt to changing environments, yet have been limited in their capacity to quantify and use adaptive genetic variation for prioritizing populations for management. Until recently, our ability to directly investigate adaptive genetic variation has been limited to a handful of model organisms studied under artificial conditions. Population genomics combines genomic technologies with population genetics theory to provide a framework for detecting genome-wide associations between segregating variation and fitness-related traits in natural populations. This approach seeks to identify loci that exhibit signatures of selection in comparative genomic scans of populations exhibiting varying phenotypes and/or relative to environmental variation.
 Our research applies the tools of population and landscape genomics to the study of adaptive divergence within systems of ecological and conservation significance, particularly kokanee salmon and American pika. Using a variety of approaches including whole genome sequencing, RADseq, and RNAseq, this work has provided basic insights into life-history evolution and the genetic basis of adaptation within natural populations at multiple spatial scales (local, regional and landscape). Importantly, this work has allowed us to develop tools, typically in the form of genotyping-by-sequencing SNP panels, that help inform wildlife and fisheries management.
Our research applies the tools of population and landscape genomics to the study of adaptive divergence within systems of ecological and conservation significance, particularly kokanee salmon and American pika. Using a variety of approaches including whole genome sequencing, RADseq, and RNAseq, this work has provided basic insights into life-history evolution and the genetic basis of adaptation within natural populations at multiple spatial scales (local, regional and landscape). Importantly, this work has allowed us to develop tools, typically in the form of genotyping-by-sequencing SNP panels, that help inform wildlife and fisheries management.
Invasion Genetics
 The devastating impacts of invasive species on native biotas have long been recognized, as documented by 18th and 19th century naturalists-explorers. Today, invasive species are considered among the most significant threats to global biodiversity, impacting native species, community structures, and entire ecosystems. Genetics and genomics offer important tools for invasive species management in order to fill several knowledge gaps including, but not limited to, sources of invasion, invasion pathways, eradication units, and detection. Work in the ECGL employs population genomics to better understand all aspects of the invasion process and to directly inform management, including for invasive rats and deer within island systems (e.g. Galápagos, Haida Gwaii) and monk parakeets throughout their native and naturalized distributions.
The devastating impacts of invasive species on native biotas have long been recognized, as documented by 18th and 19th century naturalists-explorers. Today, invasive species are considered among the most significant threats to global biodiversity, impacting native species, community structures, and entire ecosystems. Genetics and genomics offer important tools for invasive species management in order to fill several knowledge gaps including, but not limited to, sources of invasion, invasion pathways, eradication units, and detection. Work in the ECGL employs population genomics to better understand all aspects of the invasion process and to directly inform management, including for invasive rats and deer within island systems (e.g. Galápagos, Haida Gwaii) and monk parakeets throughout their native and naturalized distributions.
Conservation Units and Cryptic Diversity
 Conservation biology emerged as a crisis discipline, with the goal of applying best available science towards stemming the rate of biodiversity loss. As the resources available for protection do not match the scope of the threats, significant efforts have been directed towards developing a framework for identifying and prioritizing “units of conservation”. Work in the ECGL provides substantial theoretical and empirical contributions to this research area, demonstrated in a wide range of vertebrate species of conservation concern including Amazon parrots, painted turtles, monk parakeets, Great Basin spadefoot and others. Likewise, genetic and genomic approaches have contributed to the discovery of cryptic species diversity that, in some cases, has identified novel taxa of conservation interest. As part of an international team of scientists and managers, we have been involved in on-going research involving Galápagos tortoises, revealing cryptic diversity at risk and the “rediscovery” of species previously considered extinct.
Conservation biology emerged as a crisis discipline, with the goal of applying best available science towards stemming the rate of biodiversity loss. As the resources available for protection do not match the scope of the threats, significant efforts have been directed towards developing a framework for identifying and prioritizing “units of conservation”. Work in the ECGL provides substantial theoretical and empirical contributions to this research area, demonstrated in a wide range of vertebrate species of conservation concern including Amazon parrots, painted turtles, monk parakeets, Great Basin spadefoot and others. Likewise, genetic and genomic approaches have contributed to the discovery of cryptic species diversity that, in some cases, has identified novel taxa of conservation interest. As part of an international team of scientists and managers, we have been involved in on-going research involving Galápagos tortoises, revealing cryptic diversity at risk and the “rediscovery” of species previously considered extinct.
Ex Situ Conservation Genetics
 Biodiversity conservation strategies are most effective when directed towards protecting populations and ameliorating threats within their native habitat (i.e. in situ). Yet, in situ strategies may not be sufficient in all cases, requiring the management of populations outside of their native range (i.e. ex situ) using tools such as captive breeding, repatriation and/or head-starting. In the most extreme cases, where species are extinct in the wild and only persist in captivity, ex situ management may represent the only viable strategy for preventing their complete loss. Work in the ECGL uses genetic information to test theory underlying conventional ex situ population management and to refine strategies for maximizing founder genetic variation, equalizing founder contribution to the population’s gene pool, and minimizing fitness consequences for meeting conservation program goals. Study systems in this research area include Galápagos tortoises, Amur tigers, Lowland tapirs and Amazon parrots.
Biodiversity conservation strategies are most effective when directed towards protecting populations and ameliorating threats within their native habitat (i.e. in situ). Yet, in situ strategies may not be sufficient in all cases, requiring the management of populations outside of their native range (i.e. ex situ) using tools such as captive breeding, repatriation and/or head-starting. In the most extreme cases, where species are extinct in the wild and only persist in captivity, ex situ management may represent the only viable strategy for preventing their complete loss. Work in the ECGL uses genetic information to test theory underlying conventional ex situ population management and to refine strategies for maximizing founder genetic variation, equalizing founder contribution to the population’s gene pool, and minimizing fitness consequences for meeting conservation program goals. Study systems in this research area include Galápagos tortoises, Amur tigers, Lowland tapirs and Amazon parrots.
