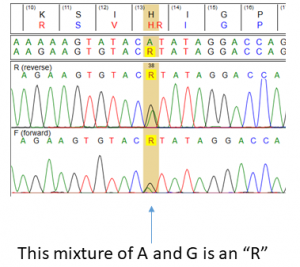
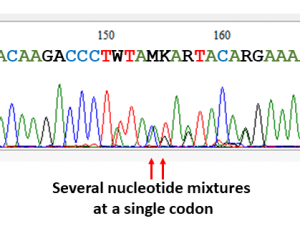
Our previous posts discussed the issue of creating “fictional” amino acids variants as a result of nucleic acid mixtures in Sanger sequences or consensus NGS sequences. Here we will discuss the OPPOSITE problem, where nucleic acid mixtures can results in the DISAPPEARANCE of actually observed mutations (and/or resistance) at when the results are submitted to online databases for processing. I am going to use examples from HIV because that is what I have been doing since I was a little boy, but the same would apply to HCV.
As we discussed below, nucleotide mixtures are indicated by IUPAC codes, such that WYC (for example, at HIV RT Codon 215) translates into four amino acids, such as Phe/Thr/Cys/Ser. Usually we would write this as T215P/T/C/S, and this is fair enough. However, by the time that one starts to get quite a few nucleotide mixtures detected at a given codon, things start to get a bit unwieldy: an NNN would translate to T215A/C/D/E/F/G/H/I/K/L/M/N/P/R/S/T/V/W/*. This would obviously be absurd to report to patients, and is also such a high multiplicity of possible mixtures at a given codon would usually be an indication that the data quality (using Sanger Sequencing) was very poor.
As a result, most people (and programs) have decided that if they see a mixture containing “too many amino acids” – probably of more than four – at a given resistance position, they would declares the position an X (which is fair enough) and INTERPRET THAT THE MUTANT AMINO ACID IS EFFECTIVELY **NOT** PRESENT. This seems to be something similar to what the major databases which report resistance have done.
This is a feature or a bug, depending on how you look at it. I suspect that the rationale was that if there were more than 4 amino acids present, it was therefore present at less than ~20% of the population and could not be detected *by Sanger Sequencing* anyway, and this makes a lot of sense. However, this can become a problem if you have a consensus sequence created from NGS data. If you have a genuine mixture of a mutations, and progressively add some minority species data (apparently “noise”, but what is actual data), it can ACTIVELY REMOVE the mutant signal….so for example, it can claim that a mutation is “present” (and a sample “resistant”) at a 20% or a 5% cut-off yet absent (and a sample sensitive) at a 2% cut-off, as a result of collapsing “too many amino acids” into an “X”. This seems to be a problem. So a lot of studies looking at different NGS cut-offs by processing consensus sequences through these databases have probably been subtracting the actual signal (this post) as well as adding to the noise (previous post) at low prevalence of mutations. The result is a systematic underestimate of the benefit of NGS data.
To demonstrate this, we looked at the effects of systematically changing only codon 184 of the HIV RT in an HXB2 sequence background with increasing amounts of different simulated types of “noise”, include the M184V (example fasta included). This mutation (and an HIV example) was chosen just because everyone universally agrees it confers resistance to 3TC/FTC, so it represents the simplest possible case. To orient you, ATG is wildtype and GTG is resistant.
We submitted the sequences to both the Stanford Website and Geno2Pheno, and the results were very interesting. Here’s the example fasta file, cleverly called “test“, if you want to play with it yourself.
Stanford Database
Base AA call Interpretation
==== ==== ========
GTG M184V resistant
STG M184LV resistant
VTG M184MLV resistant
GHM M184ADEV resistant
GHV M184ADEV resistant
GWS M184DEV resistant
BTG M184LV resistant
BMG M184X susceptible(!)
SWS M184X susceptible (!)
GNN wt (!!!) susceptible(!)
VMG M184X susceptible(!)
NNN wt(!!!) susceptible(!)
Note that A BTG at codon 184 is resistant, but BMG is susceptible. And in the case of NNN or GNN, it actively denies the mutation is even present as M184X at all. I think the most egregious offender is GNN…. if you have the key M184V mutation (the G in GTG) with too much noise AT THE OTHER TWO BASES, it declares you to be wildtype (!)
The results when processed through Geno2Pheno are different, but also also show unexpected behavior.
geno2Pheno Results
Base AA ” Interpretation
==== ==== ========
GTG M184V 3TC resistant (56-fold) (ddI, ABC partial)
STG M184V 3TC resistant (56-fold) (ddI, ABC partial)
VTG M184V 3TC resistant (56-fold) (ddI, ABC partial)
GHM M184V 3TC resistant (56-fold) (ddI, ABC partial)
GHV “missing” 3TC resistant (12-fold) (ddI partial)
GWS M184V 3TC resistant (56-fold) (ddI, ABC partial)
BTG M184V 3TC resistant (56-fold) (ddI, ABC partial)
BMG wt (!!) 3TC resistant (12-fold) (ddI partial)
SWS M184V 3TC resistant (56-fold) (ddI, ABC partial)
GNN “missing” 3TC resistant (12-fold) (ddI partial)
VHG “missing” 3TC resistant (12-fold) (ddI partial)
VMG wt (!!) 3TC resistant (12-fold) (ddI partial)
VHG “missing” 3TC resistant (12-fold) (ddI partial)
NNN “missing” 3TC resistant (12-fold) (ddI partial)
The implication of all this is that a lot of studies have probably been both subtracting the actual genuinely detected resistance signal (this post) as well as adding artificially adding to the noise at low prevalence of mutations. As a result, we should probably think again about the effect of nucleotide mixture processing on all previous studies of NGS sequencing which submitted results as consensus sequences to these databases. This is also another argument for translating NGS data before converting a consensus sequence.