Our initial few blog posts will focus on some of the “interesting” or less well-known details of using DNA sequencing methods to monitor drug resistance. A warning here that only a few obsessive people will care about these details! I’ll be mixing and matching examples from HIV and HCV.
Usually sequencing is described as something like “determining the sequence of A, C, T, and G” in the virus (after converting to DNA). But in reality the IUPAC codes are not just A,C,T and G, but also include 16 codes where DNA mixtures are observed. For example, a mixture of an A and a G, where viruses with both an A and a G are present simultaneously is denoted by an R, for “purine”. (See picture).
This is all straightforward enough. Where it gets a bit trickier is when there are two (or more) base changes in a codon, which can lead to some initially unexpected behavior. For example, one of the earliest codons of interest in HIV was position 215 of the HIV Reverse Transcriptase, commonly Threonine encoded by ACC at the nucleotide level. The nucleotides are usually ACC in most western HIV variants but when drug resistance is selected by AZT, can change to a Phenylalanine, usually TTC or a Tyrosine (usually TAC).
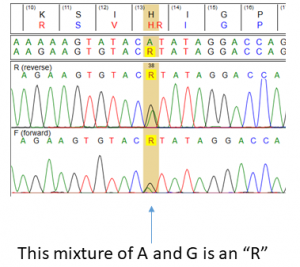
In the example above, a mixture of an A and G gives a R, which corresponds to both an Histidine and an Arginine. Now, a mixture of viruses where both drug resistant virus (“TTC”) and drug susceptible virus (“ACC”) is therefore identified by Sanger sequencing has TWO mixtures and is depicted as the nucleic acid mixtures WMC. That is great. BUT the tricky part is that when we go to translate that sequence back to amino acids, we cannot tell that there is only the starting TTC (Phe) and the ACC (Thr) present in the “WMC” nucleotide codes, so we translate this back to include two “fictional” amino acids that don’t actually exist! These are TCC (Ser) and TAC (Cys) in this case.
The result of all this is that since the 1990s we have cheerfully been reporting the existence of a lot of “drug resistant” HIV variants that literally may not exist in a given sample, including 215(Serine) and 215(Cysteine). As it turns out, this has not been a big problem, just a bit embarrassing. Similar “fictional” amino acids can happen with the translation of mixtures of other drug resistant and wild-type viruses, as long as there is a two-base change.
What can we do about this? It depends on the sequencing methods you are using. If one is using Sanger sequencing, there is really nothing one can do, short of cloning or diluting out the individual virus strains present one at a time. Life is too short for that. However, if using NGS sequencing methods, you CAN avoid creating these fictional amino acids by doing translations to amino acids BEFORE creating consensus sequences. There is a new format called AAVF which allows one to deal rationally with NGS sequences and prevents the invention of “fictional” DNA sequences.
Our next post will deal with a particularly devious example of this popping up as a potential problem for HCV analyses.
