Kanduri, C. (2016). Long non-coding RNAs: lessons from genomic imprinting. Biochimica and Biophysica Acta.
Introduction
- In gametogenesis ~1% of protein-coding genes undergo genomic imprinting
- >150 imprinted genes identified in mouse
- Typically located in clusters that range from a few kB to 3.0 Mb
- Long non-coding RNAs found in all imprinted clusters
- Inverse expression pattern relative to protein-coding counterparts
- Promoters map to differentially methylated regions (DMRs)
- Deletion of the DMRs often leads to loss of imprinting
- ICRs: imprinting control regions, 1-3 kb in size, most DMRs are ICRs
- Human genome has more lncRNAs than protein-coding genes
- Perform various functions in development, differentiation, disease
- Multiple mechanisms both at transcriptional and post-transcriptional level
- Most target chromatin modifying complexes e.g. PRC2, SW1/SNF, hnRNPK, G9a
- Implicated in gene regulation at post-transcriptional level when they are localized to the cytoplasm
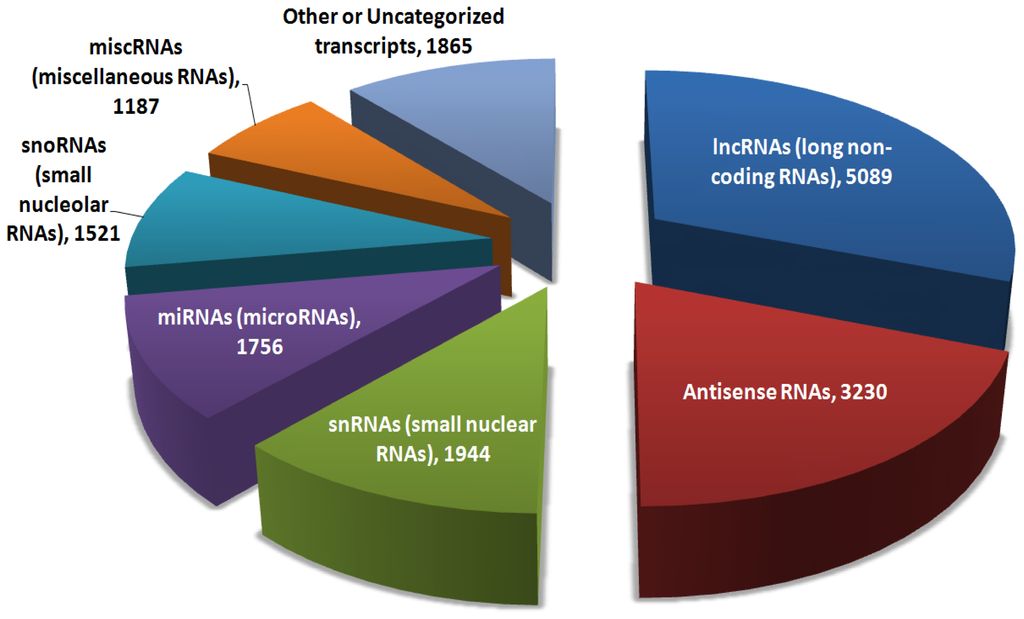
Da Sacco et al. (2012). IJMS. Pie chart of the major categories of >16,000 non-coding RNAs in the genome.
Intergenic lncRNAs in Genomic Imprinting
- lncRNAs can be classified into 4 main categories: intergenic, antisense, intronic, and enhancer
- All except intronic implicated in imprinting and parent-of-origin specific expression
- H19: lncRNA (2.3 kb in length)
- Maps to a well-investigated cluster on mouse chromosome 7, human chromosome 11
- only expressed from the maternal allele, silenced on paternal
- Paternal allele silenced via CpG methylation at the promoter
- May have lineage-specific roles in the body
- Deletion of H19 affected Igf2 in mesoderm but not endoderm
- Deletion also has effects on growth, but is not embryonic lethal
- Part of IGN (imprinted gene network) that includes 16 genes
- Potentially controls growth via regulation of the IGN in trans
- Interacts with MBD1, a methyl CpG-binding protein
- Complex recruits H3K9 methyltransferase to DMRs of some members of the gene network
- Establishes H3K9me3 marks to “fine tune” expression from both parental allelles
- Expressed at high level in embryogenesis, downregulated after birth
- Exception: remains highly expressed in muscle tissue
- May promote myogenic differentiation?
- Also shown to have oncogenic, tumour-suppressive properties
- Overexpression of H19 is linked to metastasis
- IPW: paternally expressed lncRNA, maps to an imprinting cluster on mouse chromosome 7, human chromosome 15
- Deletion observed in 70% of cases of Prader-Willi syndrome patients
- Mouse expression mainly restricted to the brain, but humans seen in all tissues
- 5′ end contains tandem repeats
- functional role of IPW cluster has not been investigated, but shown to interact with G9a methyltransferase
- Targets IG-DMR to modify chromatin structure via H3K9
- IG-DMR is a master controller of gene expression at the DLK-D103 imprinted cluster
- First example of a lncRNA shown to promote chrosstalk between two imprinting clusters by altering ICR chromatin
- MEG3: maternally expressed gene 3, imprinted lncRNA, maps to DLK-DI03 locus on human chromosome 14, mouse chromosome 12
- IG-DMR controls maternal specific expression of MEG3
- MEG3 expression is a marker of iPSCs with a fully pluripotent state
- iPSCs without MEG3 expression are not viable to support embryonic development
- MEG3 may promote interaction of PRC2 with JARID2
- Complex of PRC2-JARID2 shown to contribute to ESC differentiation
- MEG3 may therefore underlie the fully pluripotent state
Enhancer RNAs in Genomic Imprinting
- Enhancers are responsible for spatio-temporal gene regulation
- “landing site” for transcription factors, co-activator complexes
- Can activate or increase transcription from distal promoters
- Characteristics of enhancers include:
- DNase I hypersensitivity
- Post-translational histone modifications (especially H3K4me1/2, H3K27ac)
- Bidirectional transcription
- Transcripts generated are low copy number, non-polyadenylated
- Enhancers promote target gene expression via recruitment or stabilization of basic transcription machinery binding
- Establish higher-order chromatin contacts between enhancer and targets
- Transcripts from IG-DMR control expression of maternally expressed transcripts at DLK1-Dio3 locus
- Methylated on the paternal chromosome, unmethylated on the maternal
- Unmethylated version is critical for expression of multiple lncRNAs including MEG3
- Therefore acts as putative enhancer with enhancer-specific histone marks, encodes bidirectionally transcribed ncRNAs
- Maternal chromosome bidirectional transcription in ESCs correlates with early replication, inner subnuclear positioning of Dlk1-Dio3 locus
- IG-DMR transcripts may promote higher order chromatin
- Enables early replication, subnuclear localization
- Maintenance of expression of maternally expressed genes