Today is the last microbiology lab! In last week’s lab we incubated the bacteria isolated from TSI slant on API20E incubation tray for one week.
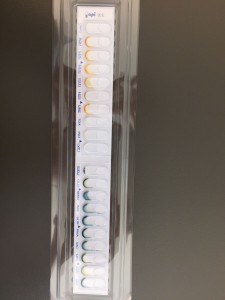
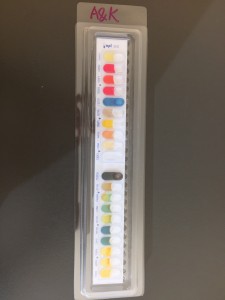
This week, we got back the API20E incubation tray and then added some reagents to observe the test results. After the observation of each test chamber, we recorded the test result as positive or negative. Then we used the API web to analysis our results. Noticed the colors of some API solutions actually changed after adding reagents. For example, the color of VP changed to red from colorless. In GLUcose, once reagent was added, it turned red immediately. For most of the results, we can confidently record it as positive or negative. However, for the test chamber MANnitol, there was a combination of color of both the positive and negative results and we didn’t have H2O2 solution to do a further test. Therefore, we recorded it with a question mark.
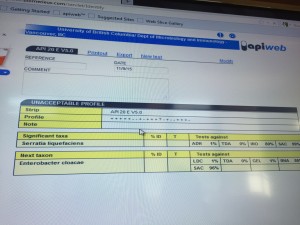
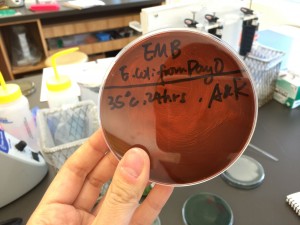
The tests results showed that it was an unacceptable profile so there’s no data corresponding to the bacteria we isolated. However, it indicated that the significant taxa is Serratia liquefactiens and the next taxon is Enterobacter cloacae.
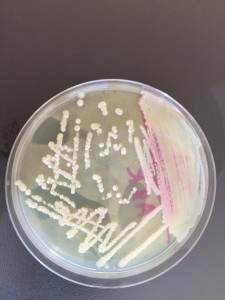
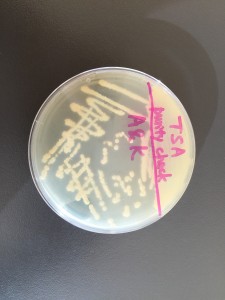
We streaked a TSA plate with the isolated colonies to do a purity check. There’s only one type of colony growing on the TSA plate so we can consider it as pure.
Thanks for keeping up with my blog! I am very pleased to share my microbiology lab experience with all of you 🙂













Recent Comments