Summary
This post explores the effects of the so-called “curse of dimensionality” on Mahalanobis distance metrics. In simulated and real data, I demonstrate that the meaning of Mahalanobis units changes with increasing dimensionality: observations recede away from any reference coordinate due to the progressive exclusion of data space from the unit sphere. The result is that Euclidean and Mahalanobis distance units do not provide a direct measure of probability density in more than one dimension. It turns out that these dimensionality effects are the essence of the statistical theory of the chi-square distribution. A meaningful distance/dissimilarity metric can be created by dividing distance by the square root of the dimensionality of the data space, which appears to create a reasonably intuitive metric based on the probability density of multivariate normal data.
Introduction
The premise of my research so far has been that the historical range of interannual climatic variability provides an ecologically meaningful metric of climatic differences over space and time. Building on the standardized Euclidean distance approach of Williams et al. (2007), I have developed a Mahalanobis metric based on the average standard deviation of historical interannual climatic variability across representative locations in any given study area. My intent is to use this metric to measure the degree to which future climatic conditions differ from those found in the current climatic classification. in other words, I intend to use the standard deviation of the historical range of variability as a unit of distance between future conditions and present conditions.
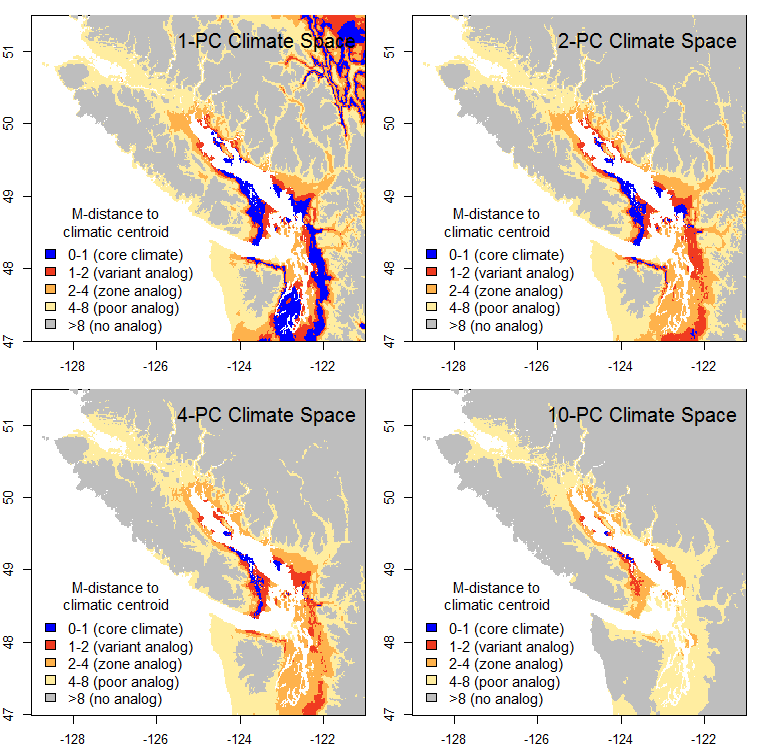
The first indication that something was missing from this approach was when I made maps of the distance from 1971-2000 normals to the 1971-2000 normals for a single location representing a BEC variant. For example, in the climatic distance map for the CDFmm (Figure 1), a very limited area is within 1 unit of the centroid, and most of the CDFmm is outside 2 units. If 1 M unit represents a standard deviation of temporal climatic variability, I would have expected most of the climate normals in the Georgia Basin to be within 2, if not 1, standard deviations from the centroid location.
Two reasons came to mind for the apparent overestimation of climatic distance: The first potential reason is overfitting. I used 10 principal components, and I standardized all the PCs to achieve a Mahalanobis metric. Given that the 10th PC is given the same weight as the first, some amount of overfitting might be expected. However, I demonstrated in my last post that overfitting due to retained dimensions was not apparent in climate year classification results. Furthermore, adding dimensions with low spatial variability should reduce a Mahalanobis dissimilarity estimate, i.e. do the opposite to what seems to be the problem. The other possible reason is the “curse of dimensionality.” I looked into it, and it turns out that my thinking about distance was neglecting some fundamental geometric realities.
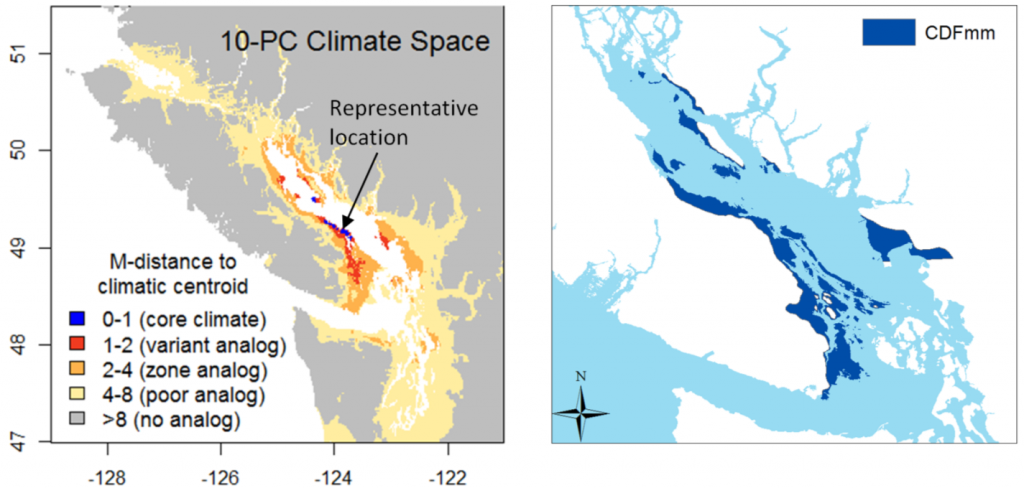
Figure 1: Mahalanobis distance from the 1970-2000 climatic normals in the Pacific Northwest to a location representing the average climate of the CDFmm.
How does the map change with different numbers of PCs?
The obvious firsts step is to see how the map changes at different levels of dimensionality (numbers of PCs retained) (Figure 2). One principal component is obviously too little information: false analogues are abundant in the interior rainshadow. The 2-PC climate space yields much better results, as would be expected because there is another key dimension of climatic variability to measure distance in. The 4 and 10-PC climate spaces show the core areas becoming progressively more distant, even though the outer limits of the range of analogues contracts only subtly. Given that my previous posts have demonstrated that the spatial variation in climate is predominantly captured in the first three PCs, this non-proportionate “distancing” phenomenon is suspect.
Another way to see the effect of dimensionality is in histograms of the distance of individual climate years to their own centroid (Figure 3). In one dimension, the distance to centroid takes the expected half-normal distribution, in which most climate years are within one M unit (i.e. one standard deviation) of their centroid, and almost all are within two M units. However, this distribution becomes altered in climate spaces of more than one dimension. The distances to centroid increase at higher dimensions, to the extent that in 10 dimensions there are no climate years within 1 M unit of the reference condition (i.e. the origin of the climate space). This indicates that the meaning of distance and probability density changes with increasing dimensionality.
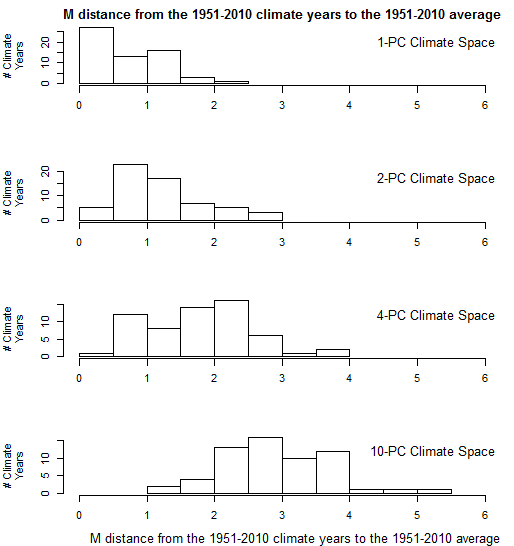
Figure 3: effect of dimensionality on the Mahalanobis distance of CDFmm 1951-2010 climate years to their own centroid.
The shift in the distribution of distances between the mean of a sample and its observations has a very simple and intuitive geometric cause. Figure 4 is a plot of the CDFmm climate years in the first two PCS. By definition, about two-thirds of climate years lie within one standard deviation of PC1 (the area shaded gray). However, in two-dimensional space, a much smaller proportion of climate years are within a distance of one M unit of the centroid, i.e. within the unit circle. It follows that in three dimensions, even fewer observations would be within the unit sphere.
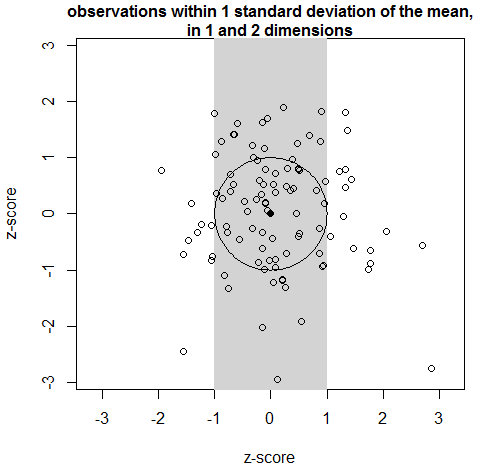
Figure 4: 2D demonstration of the reason for the exclusion of observations from the region of 1 standard deviation around the mean.
Simulations
The relationship between distance, probability density, and dimensionality can be explored through simple simulations of random data. The first simulation is simply a multivariate normal distribution with a standard deviation of one in each of i dimensions. At dimensionality higher than one, distances to centroid rapidly approach a Gaussian-like distribution with a standard deviation of 0.707 (square root of 2). This distribution is maintained at dimensionality higher than 3, even though the distance from the centroid increases. As a result, the ratio of the distance to centroid of the closest and furthest observations approaches 1 at very high dimensionality. The explicit description of this effect is generally attributed to Beyer et al. (1999).
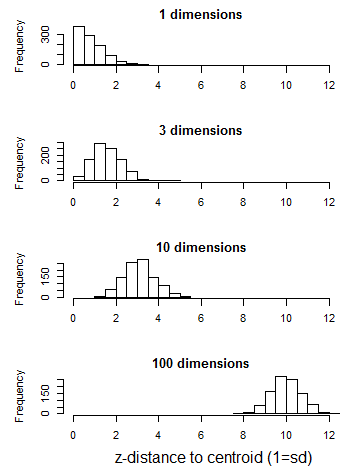
Figure 5: distance of observations of a simulated random normal sample (N=1000, sd=1) from their own centroid.
Simulation 2 explores the case of two random normal samples with the same centroid but different standard deviations (Figure 6). Although the two samples overlap in low dimensional space (<4 dimensions), they diverge from the mean (the origin) at different rates such that they are completely separate in high dimensional space. The standard deviation of distances to centroid is the standard deviation of the univariate data distribution divided by the square root of two (Figure 7). The mean distance to centroid is standard deviation of the data multiplied by the square root of the dimensionality.
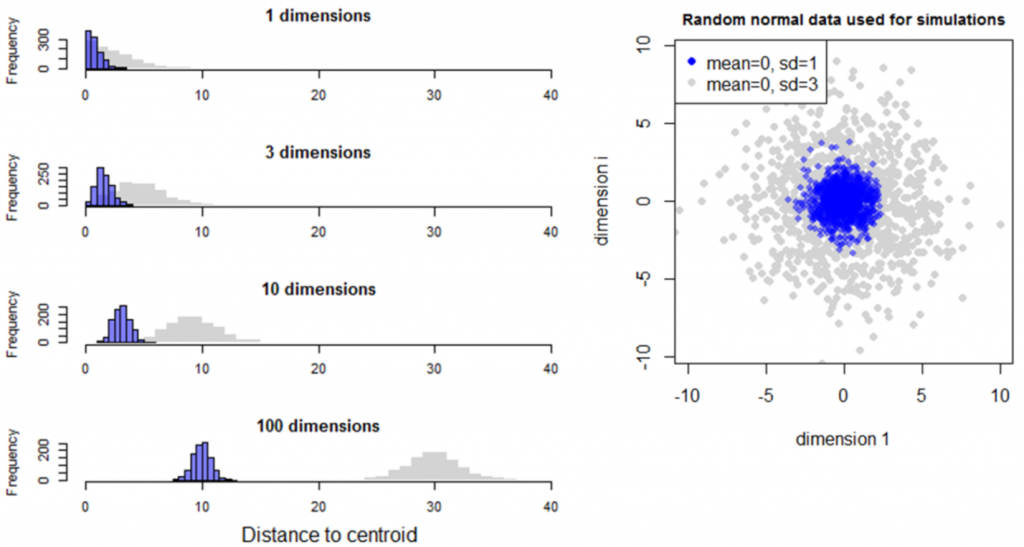
Figure 6: Simulation 2—Effect of dimensionality on distance to centroid in random multivariate normal data with the same mean but different standard deviations.
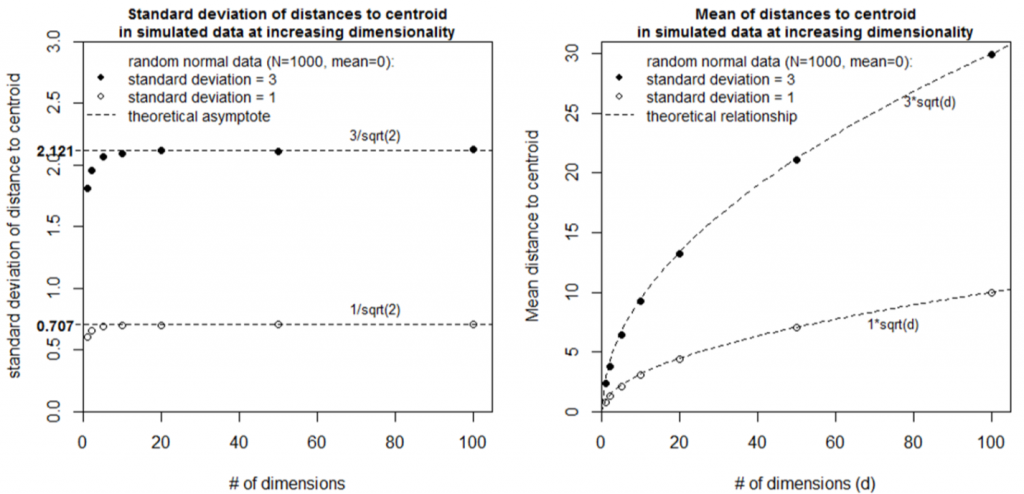
Figure 7: Simulations used to infer geometrical relationships between dimensionality and the distribution of distances to centroid.
Simulation 3 adds an additional multivariate normal sample with a standard deviation of 1 and a mean of 3 in all dimensions (Figure 8). The distance from the origin to these observations (red distribution) is slightly greater than that of the multivariate normal sample with a mean of zero and standard deviation of 3 (grey distribution), though the mathematical relationship is not immediately clear. The standard deviation of the red sample is slightly less than one (0.98) in all levels of dimensionality. This is likely because the tail of the univariate distribution of this sample crosses the origin, resulting in a slightly folded distribution of distances to the origin. Nevertheless, this simulation indicates that dispersion (standard deviation of samples) is preserved in multidimensional distances.
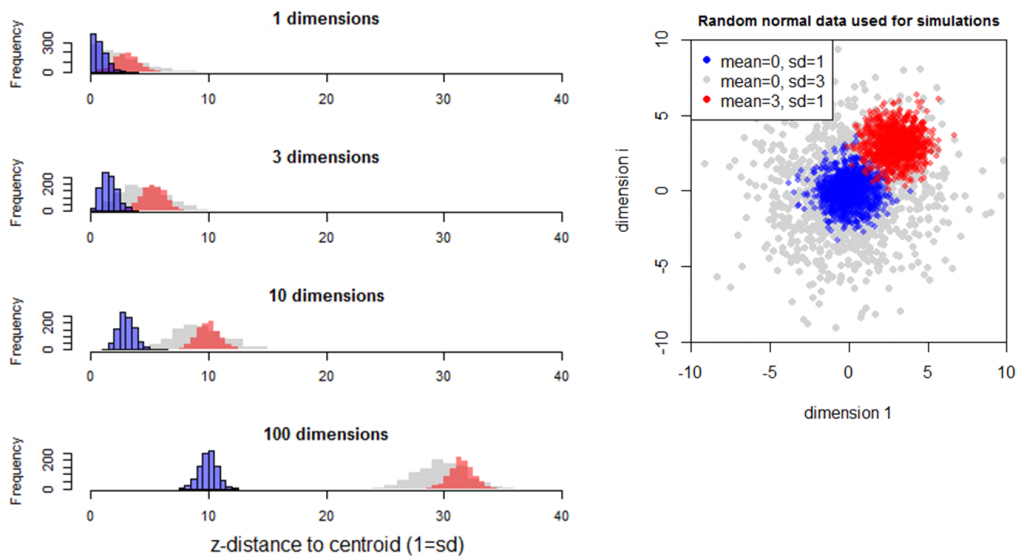
Figure 8: Simulation 3—Effect of dimensionality on distance to an additional multivariate normal sample with a standard deviation of 1 and a mean of 3 in all dimensions.
The last simulation investigates the effect of a sample mean being different in only one of many dimensions (Figure 9). In this case, the mean of distances of the different sample (red) approaches that of the reference sample (blue) in higher dimensionality. In other words, the distance between samples is influenced by the number of dimensions in which the difference between the samples occurs.
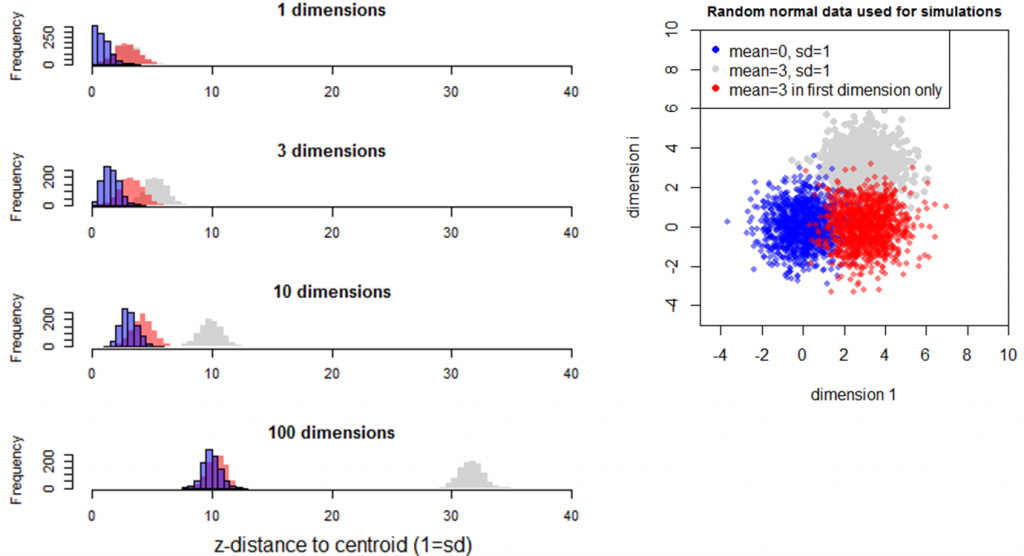
Figure 9: Simulation 4—Effect of dimensionality on distance to the origin in random samples with the same standard deviation but different means. The red sample has a mean of 3 in the first dimension, and a mean of zero in all other dimensions.
The simulations suggest the following attributes about the relationship between distance, probability density, and dimensionality:
- The relationship between distance and probability density is non-stationary under varying dimensionality. i.e. the probability of an observation occurring within a distance of one standard deviation of the sample mean decreases as the dimensionality increases.
- The probability distributions of multivariate normal data are hollow hyperspheres. The mean of the hyperspherical probability distribution is located at a distance from the centroid equaling the standard deviation of the multivariate normal data multiplied by the square root of the dimensionality. The standard deviation of the hyperspherical probability distribution is equal to the standard deviation of the multivariate normal data divided by the square root of two.
- The overlap between probability density distributions of multivariate normal data with different dispersion (i.e. different standard deviations) approaches zero with increasing dimensionality. In other words, given two different samples of the same mean but different standard deviations, the probability that observations from these two samples could occur in the same location approaches zero as dimensionality increases.
- The dispersion of probability density is preserved in high-dimensional space.
- The influence that the data distribution in any one dimension has on the total probability density of distance to centroid decreases with increasing dimensionality.
The geometrical phenomena described above are the essence of the statistical theory of the chi-square distribution. The squared Euclidean distance of standard (i.e. unit variance) multivariate normal data to their own centroid approach a chi-square distribution with degrees of freedom equalling the dimensionality of the data space (Wilks 2006). It follows that the squared Mahalanobis distance to centroid of any multivariate normal distribution will follow a chi-square distribution.
The effects of dimensionality on distance have also been studied in the data mining literature (e.g. Brin 1995, Beyer et al. 1999, Aggarwal et al. 2001), though often without recognition of the link to the chi-square distribution. In this context, they are typically referred to as an aspect of the “curse of dimensionality”, which refers to the exponential increase in data sparsity with increasing dimensionality.
Dimensionality effects are not discussed in the original formulation of the Standardized Euclidian Distance (SED) metric of novel and disappearing climates (Williams et al. 2007) nor in its regional applications (Veloz et al. 2011, Ackerly 2012, Ordonez and Williams 2013), even though dimensionality is likely an important determinant of the novelty threshold (SEDt) that is central to the metric.
Dimensionality effects in the spatiotemporal climate space of the BEC system.
How do these dimensionality effects play out in the spatiotemporal data set I am using for my analysis? A distance histogram of BC climates relative to the CDFmm variant (Figure 10) is quite revealing.
The mean of the probability distribution of CDFmm climate years is approximately the square root of the dimensionality. This would be expected, since the purpose of spatiotemporal standardization is to normalize temporal variability at each BEC variant centroid to approximate multivariate normality with a standard deviation of one. However, the distance to other BEC variants is relative stable at dimensionality more than two. The reason for this is likely because the first two PCs contain the vast majority of the spatial variation of the data (Figure 11). The other dimensions primarily represent different modes of temporal climatic variation, and therefore all variation in these lesser dimensions has approximately unit standard deviation. The result is that the relative distance between CDFmm climate years and those of other BEC variants decreases at higher dimensionality.
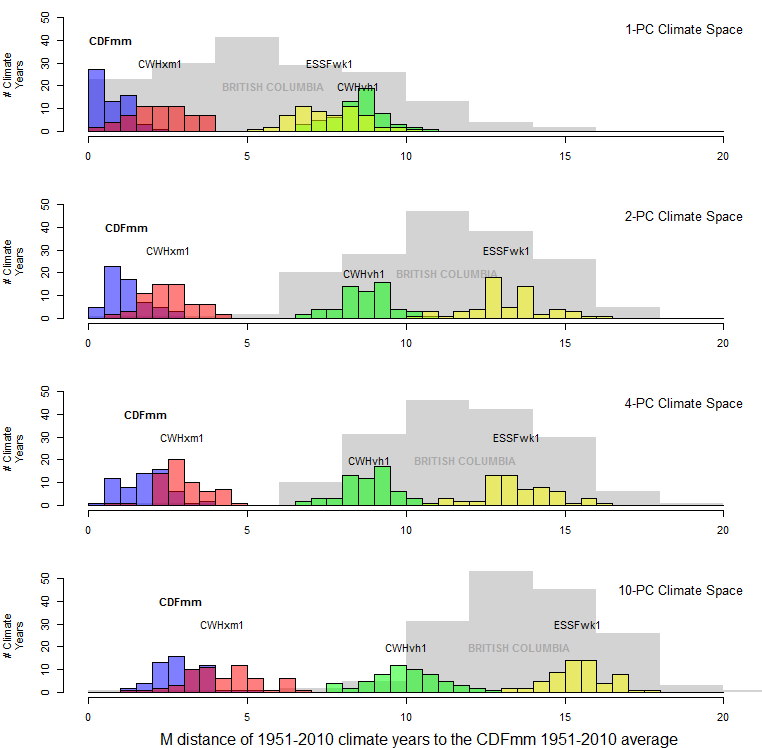
Figure 10: distance to the centroid of the CDFmm 1951-2010 climate years. Coloured histograms are 1951-2010 climate years for BEC variants representing similar (CWHxm1), somewhat different (CWHvh1), and very different (ESSFwk1) climates. The grey histogram is the distribution of 1971-2000 climate normals for 168 BEC variant representative locations.
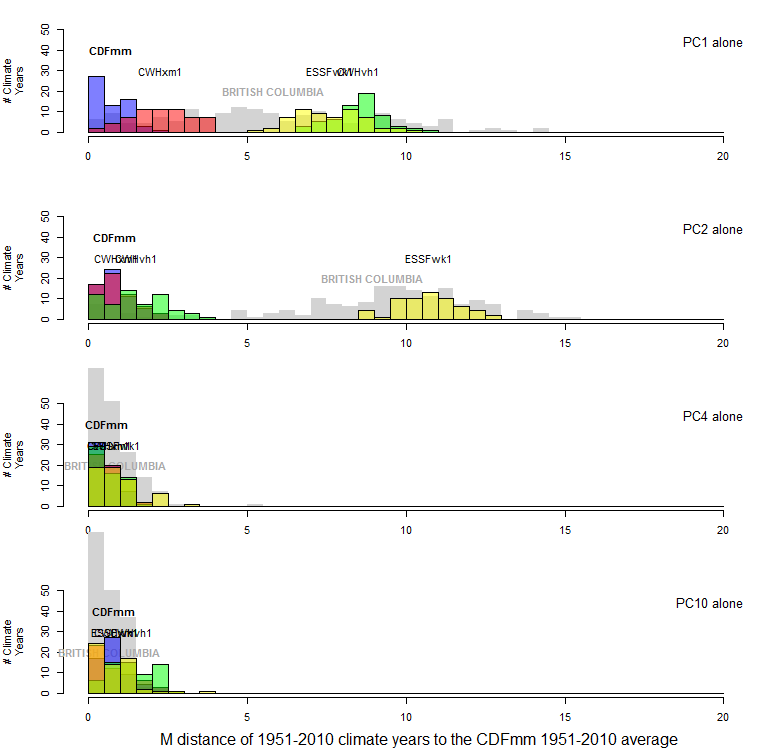
Figure 11: One-dimensional distance to the centroid of the CDFmm 1951-2010 climate years, for selected principal components. Coloured histograms are 1951-2010 climate years for BEC variants representing similar (CWHxm1), somewhat different (CWHvh1), and very different (ESSFwk1) climates. The grey histogram is the distribution of 1971-2000 climate normals for 168 BEC variant representative locations.
A solution: Dimensionality-adjusted Mahalanobis distance
My main interest in the relationship between distance, probability density, and dimensionality is in finding a good way of measuring differences between climatic conditions. My goal is to achieve a unit of distance in climate space that reflects the scale of interannual climatic variability at any given location. the problem with the dimensionality effects is that they distort the meaning of the distance metric in terms of the probability density of variability around a reference condition. One way to correct for this aspect of the problem is to standardize distances so that the probability distribution of the interannual variability in climate-year distances from the climate normal of the reference location has a mean of one at any level of dimensionality.
Spatiotemporal PCA (stPCA) creates a data space in which the reference period climate years at selected locations follow an approximately standard multivariate normal distribution, assuming normality of the raw variables. As a result, the squared distances of the climate years to their average (the reference normal) follow a chi-square distribution with degrees of freedom equalling the dimensionality of the data space. The mean of the standard chi-square distribution equals the degrees of freedom and thus the dimensionality. Hence, dividing distances by the square root of the dimensionality will center the climate years for any location at a value of one. Further, the units of this dimensionality adjusted distance metric are interpretable as the standard deviations of spherical multivariate normal distributions. Compared to unadjusted distance, this metric appears to be more easily interpreted because it is intuitively related to univariate dispersion.
Figure 12 illustrates the effect of standardizing distances in climate space by the square root of the dimensionality. This standardization has the desirable effect of limiting the probability distribution of the climate years for the focal geographic location (the CDFmm in this case) to a mean of one. It also has the effect of compressing the distributions of other BEC variants as dimensionality increases. This compression is a result of there being little spatial variation in dimensions above the third PC (Figure 11), i.e. all BEC variants have the same mean in these lesser eigenvectors. Hence the average dispersion of the data declines with increasing dimensionality. Adding dimensions for which all group means are the same will make the groups appear more similar, and thus reduce the distances between them. This is a much more intuitive result than the unadjusted Mahalanobis distance.
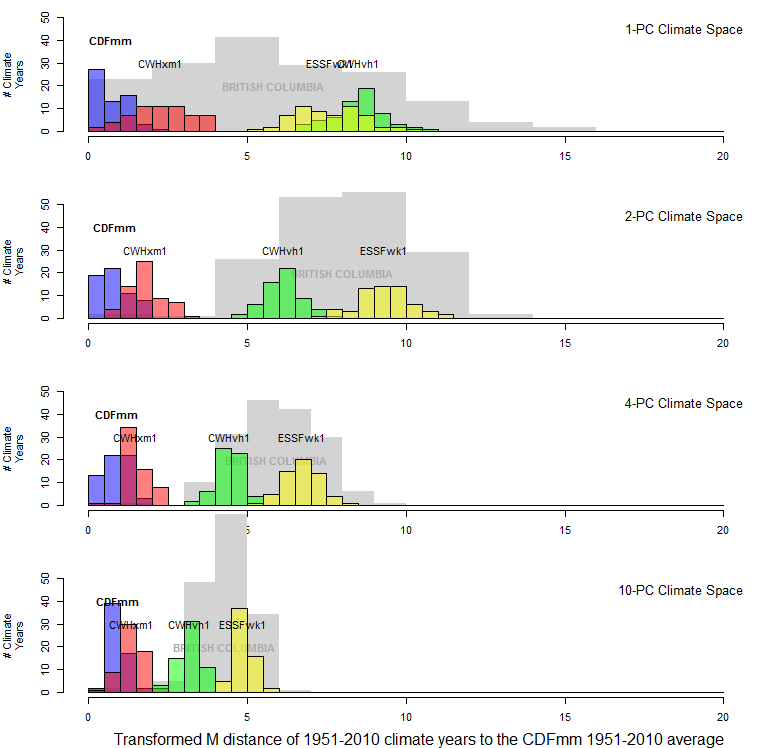
Figure 12: Effect of transforming distances to the CDFmm centroid by dividing them by the square root of the dimensionality. This transforms the Mahalanobis metric into units of standard deviations of multivariate normal data
Chi-square percentiles as an alternate distance metric
Since squared distances of climate years to their own average are expected to follow a chi-square distribution, it is logical that climatic dissimilarity could be measured in chi-square percentiles. This approach is demonstrated in Figure 13. Chi-square percentiles clearly provide a statistically precise distance metric. However, the horizon of the metric is very close to the origin: even a climate as similar to the CDFmm as the CWHdm is beyond the 99.99th percentile. This likely limits the utility of chi-square percentiles for climate change analysis.
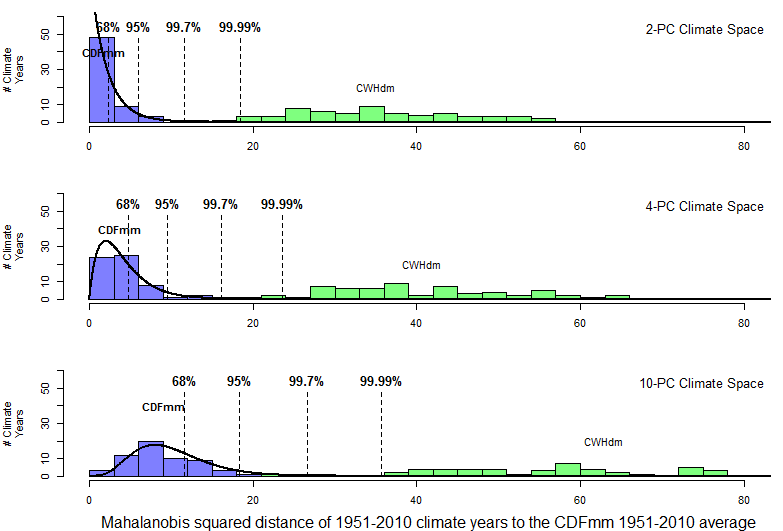
Figure 13: Utility of chi-square percentiles as a dimensionality-independent distance metric. The squared Mahalanobis distances of climate years to their own centroid follow a chi-square distribution, the percentiles of which provide a statistically precise distance metric. The horizon of this distance metric, however, is very short.
Distinction between distances to climate year distributions vs climate normals
When using the dimensionality-adjusted Mahalanobis distance as a dissimilarity metric, it is important to be mindful of the distinction between distances to climate year distributions vs climate normals. This is the distinction between the mean distance to a set of observations vs. the distance to the mean of those observations. As the mean of a standard multivariate normal distribution approaches a reference point, the distance between these two points will obviously approach zero. However, the mean dimensionality-adjusted Mahalanobis distance from any reference point to standard multivariate normal data can never be less than one, because of the inherent dispersion of the data (Figure 14). It follows from this simple logic that a distance of one has different meanings for climate year distributions vs. climate normals.
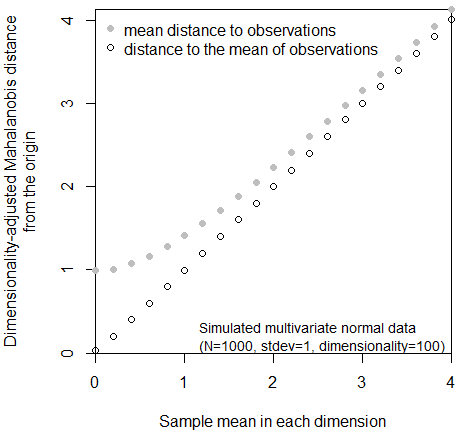
Figure 14: distinction between the mean distance to observations in a sample, as opposed to the distance to the mean (centroid) of the sample. This relationship is independent of dimensionality.
References
Ackerly, D. D. 2012. Future Climate Scenarios for California: Freezing Isoclines, Novel Climates, and Climatic Resilience of California’s Protected Areas. Page 64.
Aggarwal, C. C., A. Hinneburg, and D. A. Keim. 2001. On the Surprising Behavior of Distance Metrics in High Dimensional Space. Pages 420–434 Lecture Notes in Computer Science.
Beyer, K., J. Goldstein, R. Ramakrishnan, and U. Shaft. 1999. When is “Nearest Neighbour” Meaningful? Int. Conf. on Database Theory.
Brin, S. 1995. Near Neighbor Search in Large Metric Spaces. Pages 574–584 Proceedings of the 21st VLDB Conference. Zurich, Switzerland.
Ordonez, A., and J. W. Williams. 2013. Projected climate reshuffling based on multivariate climate-availability, climate-analog, and climate-velocity analyses: implications for community disaggregation. Climatic Change 119:659–675.
Veloz, S., J. W. Williams, D. Lorenz, M. Notaro, S. Vavrus, and D. J. Vimont. 2011. Identifying climatic analogs for Wisconsin under 21st-century climate-change scenarios. Climatic Change 112:1037–1058.
Wilks, D. S. 2006. Statistical Methods in the Atmospheric Sciences, Second Edition. Page 627. Internatio. Academic Press.
Williams, J. W., S. T. Jackson, and J. E. Kutzbach. 2007. Projected distributions of novel and disappearing climates by 2100 AD. Proceedings of the National Academy of Sciences of the United States of America 104:5738–42.