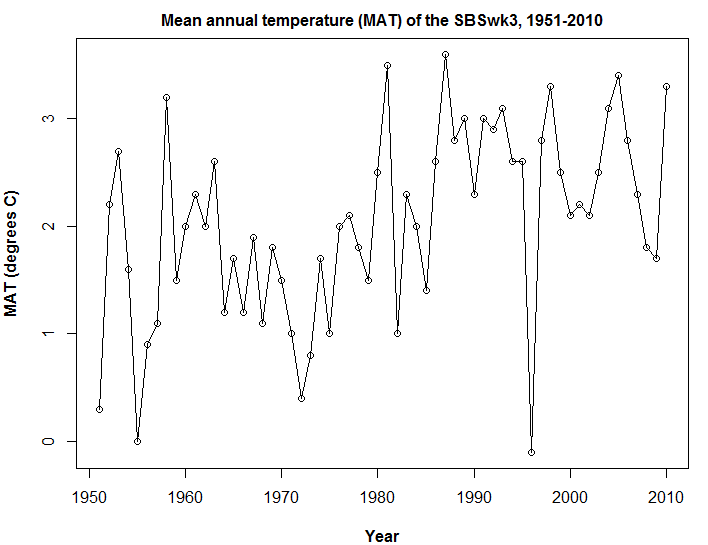
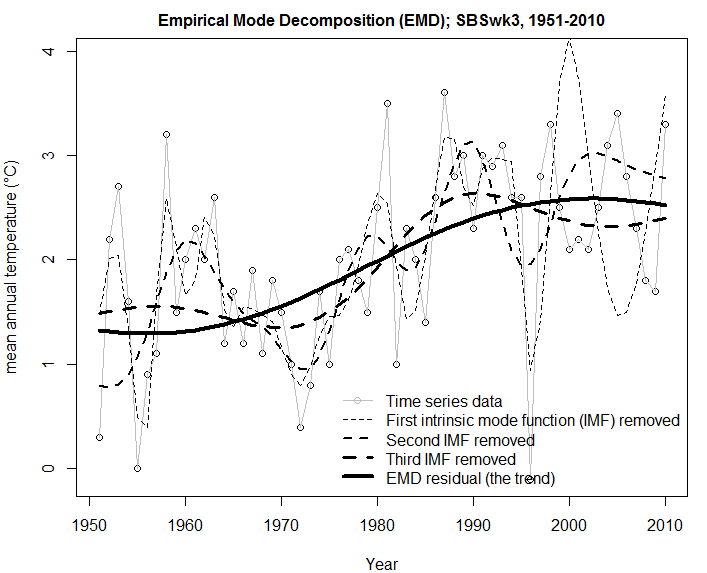
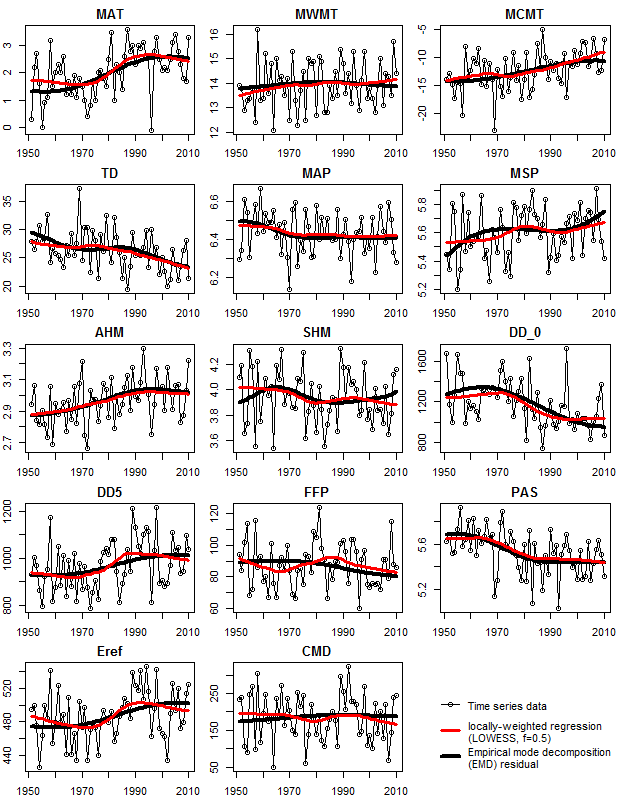
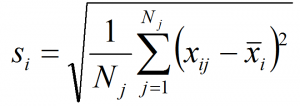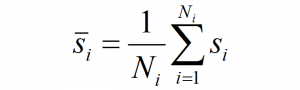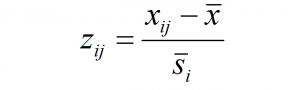Summary
A meaningful metric of climatic difference is helpful for climate change analysis. There is a strong statistical and ecological rationale for the use of Mahalanobis metrics in multivariate analysis of climatic novelties, since it removes the effect of correlations and trivial redundancies between climate variables. This post describes standardization of eigenvectors on the temporal component of spatiotemporal data. In other words, this method performs a second round of spatiotemporal standardization on features extracted from spatiotemporally standardized data. This second standardization approach appears to be functionally (and possibly mathematically) equivalent to spherical normalization of the within-groups covariance matrix of extracted features, which is commonly used within linear discriminant analysis to improve classification. In addition to its advantages in classification, this method provides a Mahalanobis metric whose units reflect the average temporal variability of the study area. This “M-metric” is a robust and ecologically meaningful metric for quantification of climatic differentiation and change.
The rationale for standardization of extracted features
Standardization (a.k.a. scaling) is a very common procedure in multivariate analysis. The basic rationale for standardization is that different variables have different and incomparable units of measurement (e.g. oC vs. mm). However, even for variables with comparable units, there is an additional ecological rationale for standardization of interannual variability: life strategies of organisms will generally be adapted to the background variability of any given climate element. Adaptation to historical variability is expected to be both temporal (e.g. winter vs. summer temperatures) and spatial (e.g. mean annual temperature in temperate vs. tropical regions) (Mora et al. 2013). This ecological rationale for standardization must be considered in metrics for climatic differences in both space and time.
Standardization gives all variables equal or comparable variance, and thus equal a priori influence on the outcome of a principle components analysis (PCA). However, PCA aggregates the correlated variance of the raw variables into the extracted principal components. The relative variance of the principal components thus reflects the redundant variance in the raw variables. Addition of a trivially and perfectly redundant variable to the raw data (e.g. a second set of MAT values), will have the non-trivial result of increasing the variance of one of the principal components and possibly increasing its rank. This behaviour highlights the importance of careful variable selection prior to performing PCA. Nevertheless, correlation between raw climate variables is inevitable, and so the ecological meaning of the unequal variance of principal components needs to be considered.
Extracted features (e.g. principal components) represent independent (uncorrelated) dimensions of climatic variation. Regardless of whether unequal variance in the principal components is due to trivial redundancies or meaningful correlations, the relative scales of variation in the principal components are unlikely to be ecologically meaningful due to life strategy adaptation. What is important is the magnitude of a climatic difference or change relative to the background variation of any given principal component. In this light, the ecological rationale for standardization of raw variables also applies to principal components and other eigenvectors extracted by other methods, such as LDA.
Mahalanobis distance vs. Euclidian distance
I have demonstrated in previous posts that principal components analysis doesn’t affect the distance between observations in climate space. Therefore, Euclidian distance on non-truncated principal components is the same as Euclidian distance on raw variables. Standardization of the principal components (dividing by the standard deviation of z-scores) creates a climate space in which Euclidean distance is equivalent to Mahalanobis distance in the standardized but unrotated climate space (Stephenson 1997). The units of standardized eigenvectors (e.g. standardized principal components) therefore are Mahalanobis metrics. Similar to z-scores, the precise meaning of a Mahalanobis metric depends on how the eigenvectors are derived from the raw data (e.g. PCA vs. LDA) and how the standardization population is defined (e.g. global vs. spatiotemporal).
Spherization of temporal climate envelopes
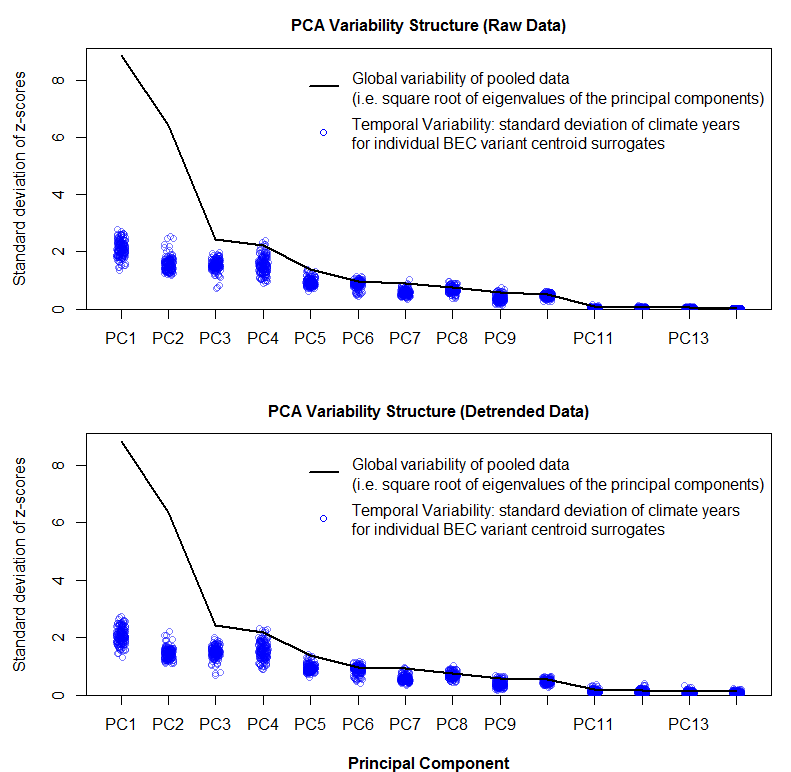
As shown in the previous post, spatiotemporal standardization operates by dividing each observation by the average standard deviation of climatic anomalies across the study area. This approach creates temporal climatic envelopes that are approximately spherical, and collectively have an average standard deviation of one z-score. Climate envelopes of individual locations will be larger or smaller, and more-or-less spherical, depending on their variability in each climate element relative to the study area average. Principal components analysis of this space is desirable, since it aggregates redundant variation into uncorrelated dimensions of variation. However, the aggregation of variance into principal components creates ellipsoidal (oblong) temporal climate envelopes. As discussed above, the ecological meaning ellipsoidal shapes of temporal climate envelope is ambiguous. A second round of spatiotemporal standardization of the principal components returns the temporal climate envelopes to an approximately spherical shape. This second standardization can be called “spherization of within-group variability,” or just “spherization” for simplicity. The metric of the sphered climate space can be called the M-score, because it represents a unit of Mahalanobis distance based on the average standard deviation of temporal anomalies of z-scores over all locations in the study area. The process of spherization of truncated principal components is illustrated in Figure 1.

Figure 1: effect of spatiotemporal standardization of principal components (spherization) on the variability structure of the BEC variants of the south-central interior BC.

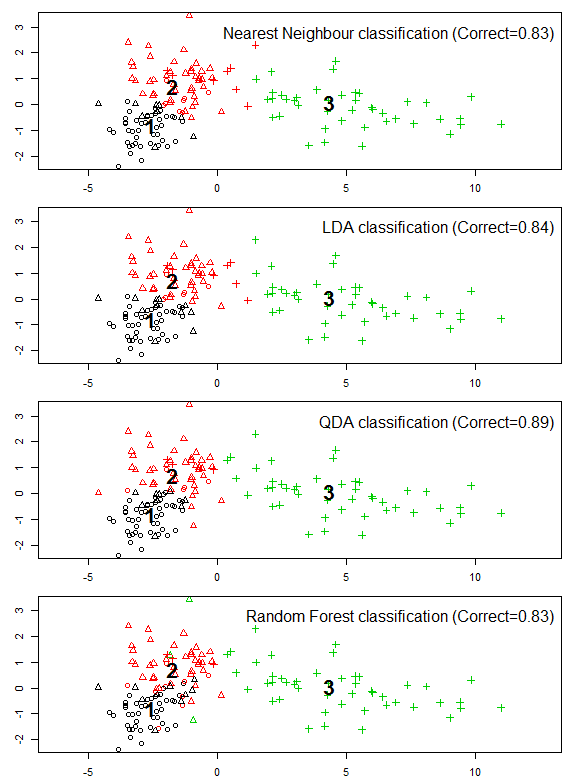
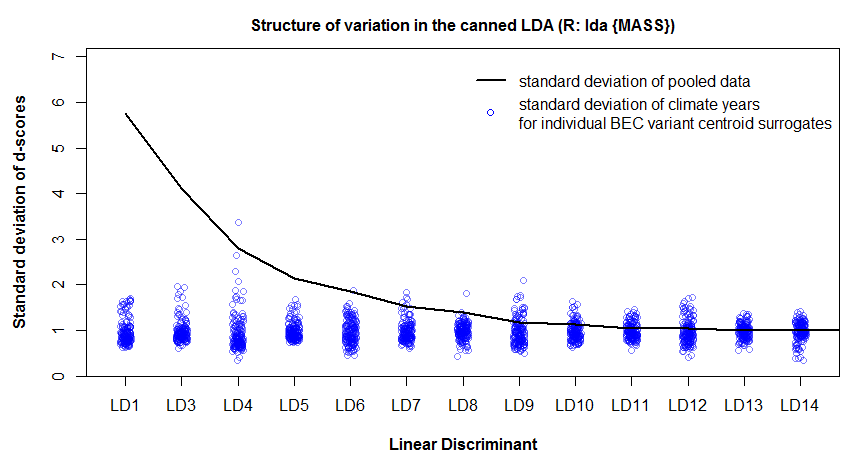
Figure 2: the role of spherization in linear discriminant analysis of the BEC variants of the south-central interior BC.
Comparison with linear discriminant analysis
Many aspects of the above procedure (spherization of local anomalies (within-group variability) of the principal components of spatiotemporally standardized data) are conceptually similar to the procedures of linear discriminant analysis. A comparison of the methods and results of the two procedures is warranted.
Spatiotemporal standardization expresses spatial variability between locations in terms of the average temporal variability of all locations. This is similar, though not equivalent, to the ratio of between-groups to within-groups scatter matrix that is the basis of LDA: The within-groups scatter matrix is the sum of the individual group covariance matrices, which is equivalent to the average covariance matrix, which in turn is likely equivalent in ratio form to the average matrix of standard deviations. Variables that have higher spatial variability will have larger variance in a spatiotemporally standardized data set. A principal components analysis of spatiotemporally standardized data is therefore functionally similar to LDA, because both give high priority (eigenvalues) to variables with a high ratio of between-group to within-group variability.
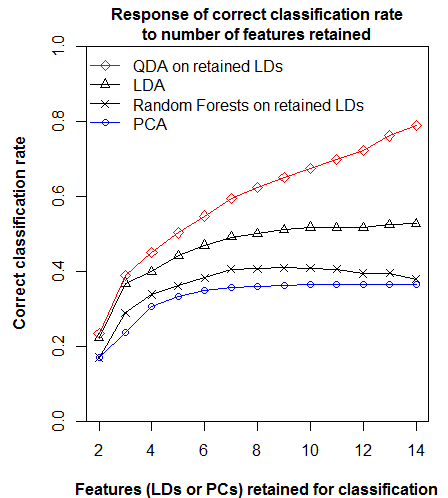
Despite this functional similarity, PCA calculates eigenvalues of the total data variance, while LDA calculates eigenvalues of the ratios of between- to within-group variance. The extracted features are different as a result. Low-eigenvalues in PCA indicate residual dimensions with very low spatial and temporal variation (Figure 1). Low eigenvalues in LDA indicate dimensions with low spatial variation, but which may nevertheless have substantial temporal variability (Figure 2). PCA is advantageous because it facilitates truncation of the features with very low variance; in Figure 1 it is clear that the last four principal components can be dropped from further analysis with minimal loss of information. However, beyond the process of truncation, the advantages of PCA vs. LDA are ambiguous. PCA structures the climate space in order of decreasing total variance, while LDA structures the climate space in order of decreasing spatial differentiation. Both of these structures are climatically and ecologically meaningful in their own way, and their relative advantages will depend on the specific research question being asked.
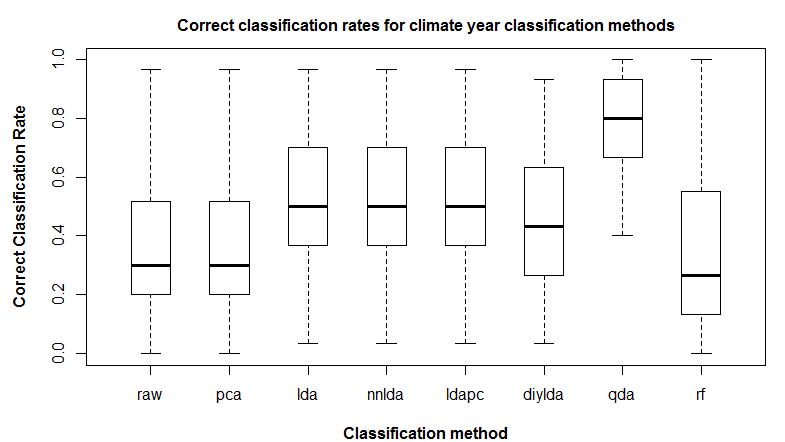
Effect of spherization on climate year classification
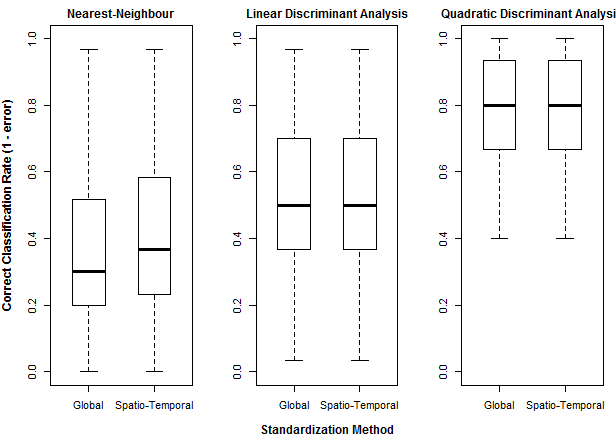
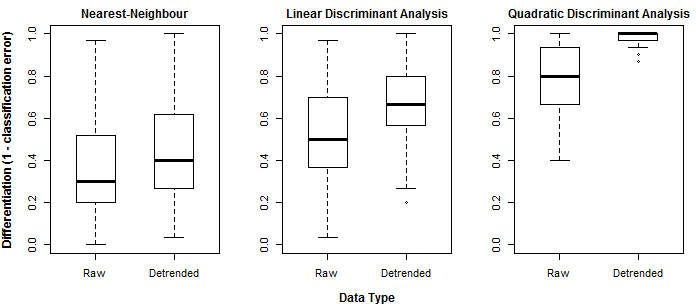
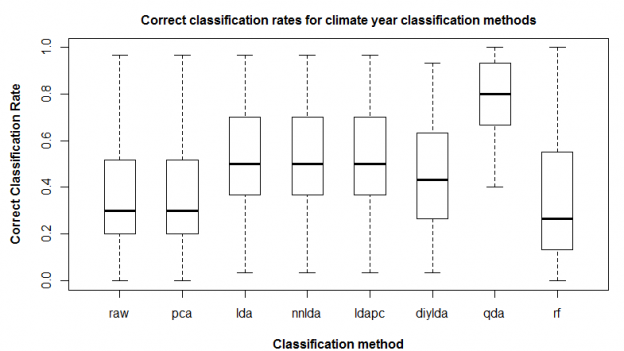
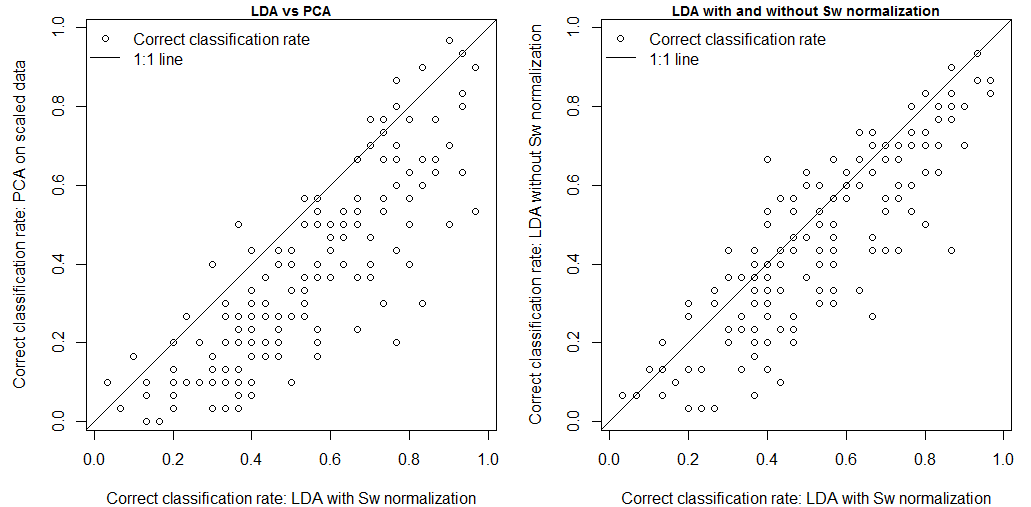
Spherization is not integral to the feature extraction component of the LDA procedure, but it is typically incorporated into LDA because it has an important role in classification. For example, it is an integral component of the lda{MASS} call in R. it is logical that spherization of the principal components should also improve nearest neighbour classification. Figure 3 demonstrates the sequential effects of spatiotemporal standardization and spherization on differentiation (nearest-neighbour correct classification rates) between BEC variant temporal climate envelopes. Typical global z-standardization produces an average differentiation in the otherwise untransformed climate space of 36%. Spatiotemporal standardization improves differentiation to 41%. As demonstrated previously, PCA has no effect on differentiation. However, spherization of the PCA improves differentiation to 47%. LDA, which includes spherization as part of its classification procedure, achieves a differentiation of 53%. It is notable that LDA without spherization (not shown in Figure 3) only achieves 44% differentiation. Spherization is clearly advantageous for classification. Spatiotemporal standardization followed by spherization of principal components achieves classification results that are only marginally poorer than LDA.

Figure 3: effect of spatiotemporal (ST) standardization and spherization on climate year classification of BEC variants, compared to simple “global” z-standardization (left) and linear discriminant analysis (LDA, right). All classifications use the nearest-neighbour criterion.
Use of spherization and Mahalanobis distance in the ecological literature climate change
Standardized anomalies are frequently used in the atmospheric sciences , and are useful for univariate analysis because they quantify variability in terms of standard deviations (z-scores). Mora et al. (2013) adapted the concept of standardized anomalies to describe “climate departures”, i.e. the year at which the future temporal climate envelope of any given location ceases to overlap with its own historical temporal climate envelope. Although they did univariate analysis on seven climate variables, they did not perform any multivariate analysis. This is very likely due to the purpose of their paper as a simple way to communicate climate change impacts to a large non-expert audience. Nevertheless, the high impact of this paper suggests that an equivalent multivariate approach is in demand.
Williams et al. (2007) provided a multivariate analysis of standardized anomalies, using a standardized Euclidean distance metric. The authors may have opted against a Mahalanobis metric for the sake of simplicity and ease of communication. Nevertheless, standardized Euclidean distance metrics are susceptible to the unjustified influence of correlations in the input variables, and so are problematic for quantitative investigation of climate change.
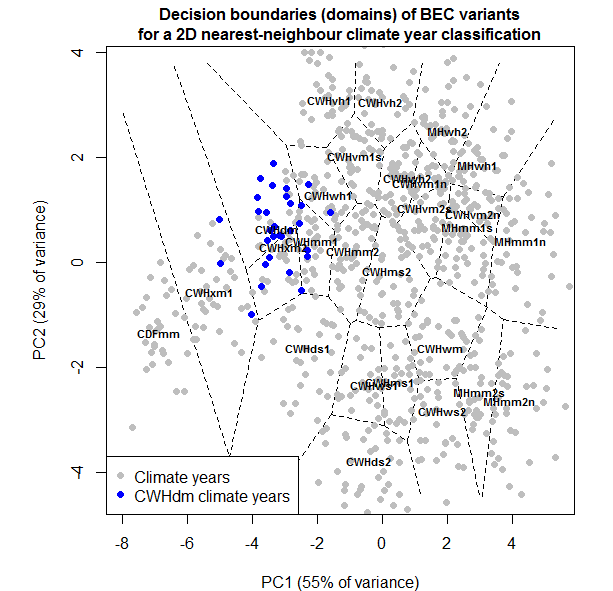
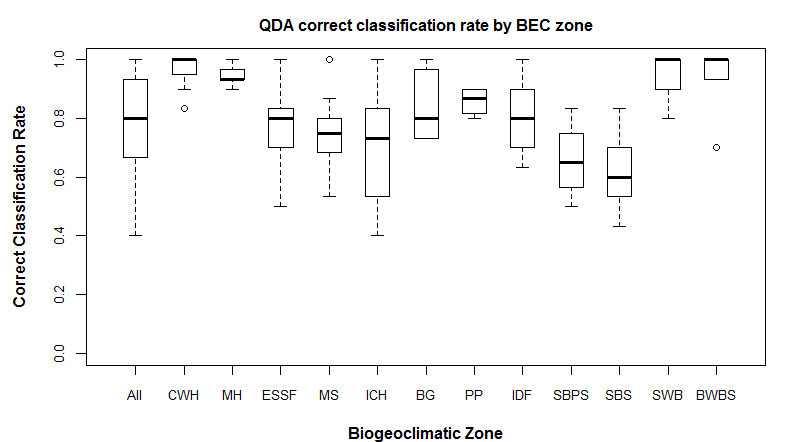
Hamann and Wang (2006) used linear discriminant analysis on spatial variation in climate normals to define the climate envelopes of BEC variants for the purposes of climate change analysis. They explicitly noted that this method uses a Mahalanobis distance for climatic classification. This Mahalanobis metric is very similar to the one I have proposed because it is based on spherization of within-group variability. However, the units of the Mahalanobis metric in Hamann and Wang (2006) reflect the average spatial variation of climate normals within BEC variants, and thus the scale of BEC mapping. A Mahalanobis metric based on the temporal variation at individual locations is essentially independent of BEC mapping, which is likely preferable for subsequent analysis of BEC mapping.
As an alternative to discriminant analysis for species distribution modeling, Roberts and Hamann (2012) proposed nearest-neighbour classification using Euclidean distance of heavily truncated principal components. They claimed that this is a modified Mahalanobis distance, but do not specify any standardization of the principal components in their methodology. Euclidian distance on principal components is the same as Euclidian distance on raw variables. As a result, it does not appear that their method produces a Mahalanobis metric, though this needs to be verified.
References
Hamann, A., and T. Wang. 2006. Potential Effects of Climate Change on Ecosystem and Tree Species Distribution in British Columbia. Ecology 87:2773–2786.
Mora, C., A. G. Frazier, R. J. Longman, R. S. Dacks, M. M. Walton, E. J. Tong, J. J. Sanchez, L. R. Kaiser, Y. O. Stender, J. M. Anderson, C. M. Ambrosino, I. Fernandez-Silva, L. M. Giuseffi, and T. W. Giambelluca. 2013. The projected timing of climate departure from recent variability. Nature 502:183–7.
Roberts, D. R., and A. Hamann. 2012. Method selection for species distribution modelling: are temporally or spatially independent evaluations necessary? Ecography 35:792–802.
Stephenson, D. B. 1997. Correlation of spatial climate/weather maps and the advantages of using the Mahalanobis metric in predictions. Tellus 49A:513–527.
Williams, J. W., S. T. Jackson, and J. E. Kutzbach. 2007. Projected distributions of novel and disappearing climates by 2100 AD. Proceedings of the National Academy of Sciences of the United States of America 104:5738–42.