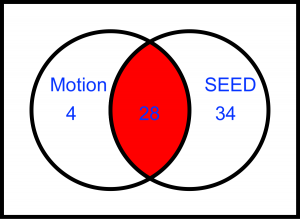
Goal is to merge two datasets, Motion and SEED. due to different dimensions I have to drop 34 channels from SEED and use only 28 channels for input.
Power-Spectral Density Function

the EEG signals are a time series data stream, the above figure shows the power spectral density of the 28 channels, this function uses Fourier transform to convert time domain to frequency domain. This allows us to learn about relationships that are not sequential like the weather patterns but like population growth patterns (refer to the hare and lynx population problem).
28 EEG Sensor Placement
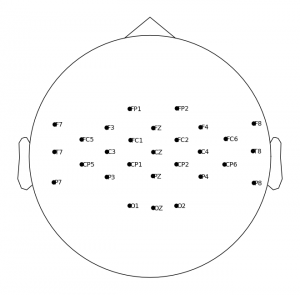
the sensor placement above shows the 28 channels that are present in both datasets.
There are 3 methods that seem feasible to go ahead with to achieve our goal of using EEG signals to send distress calls in need.
1. Separate Features -> Ensemble
This method is quite extensive to code, but the logic is simple and would most likely provide the best results.
we use a total of 56 features (EEG channels), 28 from the intersection and the other 28 being a weighted sample from the Motion and Seed sets. To train several CNN models for every combination of 56 features.
The weights would be calculated using gradients, similar groups of EEG would be selected based on gradients. this would help us identify nodes that have similar patterns with a given motion and emotion.
In the end we would have an ensemble layer, that compiles the best performers of all the networks trained, from this layer we would be able to identify the important electrode nodes to use to send distress signals.
2. Downsample
Is our Minimum Viable Product, this is the easiest model to implement, as we restrict our dataset to 28 channels for input and then to overcome different dimensions of the arrays we vary the batch size accordingly for a larger sample we would use smaller batch size and vice versa.
There are various optimization techniques we can implement in this method. such as:
- alternating strides and padding to improve training.
- Spatial Decomposition Analysis
- Time Series Techniques such as LSTM, (which is essentially telling our network layers to retain information about t-1, while optimizing the loss function at t)
- Attention factors ( allow the network to decide and accordingly assign weights to nodes that are not false/noisy)
3. NeuroPsychology
this method is likely to be the golden solution with far greater challenges on its path. It requires extensive knowledge of neuroscience to engineer features that show causal effects rather than correlated features.
For instance there must be some channels that provide signals for Flight/Fight response in human beings, we would pool such nodes or use a linear combination of them to effectively obtain signals to such responses. This would reduce our false positives and improve our ability to save a human life.
Reference
As I dwell further in modeling, I often look back and try to understand the data as much as I can. I don’t have any experience in neuroscience, but I do have extensive skills and tools in Causal Inference, Machine Learning and Time Series Analysis.
I found the paper on learning CNN features from DE features for emotion recognition quite useful and do plan to apply similar feature engineering techniques in the motion dataset, to see if I achieve better results for a shorter run time.
Fig 1
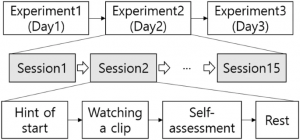
The acquisition protocol for SEED dataset
SEED dataset is well designed for EEG-based emotion classification task by machine learning or deep learning approaches. The EEG signals are recorded using an ESI NeuroScan System during watching the fifteen different emotional movie clips (five clips x three emotion labels) that enhance the emotion of participants of the dataset recording.
The dataset contains 62-channel (electrodes) EEG emotional signals which are recorded by fifteen participants (7 males and 8 females). As shown in Fig. 1, the recording of each participant has been repeated for three days, one experiment for a day, with an interval of a few days. In one experiment, there are fifteen sessions, where each session contains one emotional video clip. On the other hand, when the same stimuli are repeated to the participants, the neural activities are easily reduced .
Fig 2
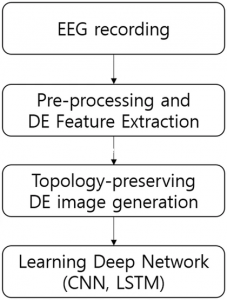
Proposed Method
Our goal is learning subject-dependant CNN-based model to recognize EEG signals by generating strong EEG features according to the proposed framework as shown in Fig. 2. We first extract DE features from recorded EEG signals, and we generate DE images which are topology-preserving DE features inspired by several previous works to learn the CNN-based model. Then, we train the proposed CNN architecture and evaluate the performance of subject-dependent emotion recognition model on SEED dataset. Optionally, to learn the temporal information of EEG signals, we learn the long–short-term memory model (LSTM).
Table 2
| Band | Frequency (Hz) |
|---|---|
| Delta | 1–3 |
| Theta | 4–7 |
| Alpha | 8–13 |
| Beta | 14–30 |
| Gamma | 31–50 |
Feature extraction
Fourier transform is often used to transform EEG signals from the time domain into the frequency domain. EEG signals are well analyzed by short time Fourier transform (STFT) as the signals are nonstationary. For EEG signal transformation, we compute STFT, and the frequency bands are selected as described in Table 2. To be conducted in the same experimental environment as in [10], we apply 256-point STFT without overlapping Hanning window of one second.
For each frequency band, we extract differential entropy (DE) as introduced in several works. DE generates stable features, which achieved high performance of EEG signal analysis [10], while EEG signals are subject to Gaussian distribution .DE can be simply defined as below [12]:
Dimensions of DE features are M×N per second (one sample), where M is the number of electrodes and N is the number of frequency bands. We filtered out redundancy components of emotional states by linear dynamic system (LDS) [41]
Fig 3
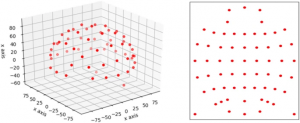
3D location of EEG electrodes (left) for 62 channels and projected 2D locations (right) using polar projection on SEED dataset
References
Learning CNN features from DE features for EEG-based emotion recognition
