There’s one thing that every single one of us has in common with each other and all other living beings. It’s the fact that we carry DNA in our cells. DNA is an important molecule that has all the inherited information about how a living thing will look and function. A strand of DNA is like an extremely long sentence that uses only four letters – in fact, the length of the human DNA is 600 billion letters long! Can you imagine how hard and time consuming it would be to read? That is why scientists at The University of British Columbia (UBC) have developed a new and exciting application called easy Visualization and Inference Toolbox for Transcriptome Analysis, or eVITTA for short. eVITTA simplifies analyzing RNA (a type of DNA) and not only makes the process more efficient, but improves scientists’ understanding of its information as well.
The kind of DNA information that eVITTA works with is called the “transcriptome”. The transcriptome is the full set of RNA, which are the copies of DNA, within a cell. Analyzing the information in the transcriptome is crucial; for the past few decades, it has been one of the most used techniques for investigating diseases and their mechanisms.
The analysis of transcriptome data, however, is very tedious and time consuming. You have to retrieve the data, examine it, and then compare it with other transcriptome datasets to draw a conclusion. This involves several steps and different types of programs, which can be inefficient. eVITTA was created to combine the many steps of transcriptome analysis into one simple, user-friendly interface. This speeds up the process of transcriptome analysis immensely!

The tedious process of analyzing long strands of DNA is simplified with eVITTA.
Image Credit: CI Photos/Shutterstock.com
evitta was born out of excess data!
One of the many topics that sparked our interest was understanding the circumstances surrounding the creation of eVITTA. As Dr. Yan of UBC’s Taubert Lab puts it:
This whole project was born out of our need to pass on data. We have a lot of transcriptome data from past years that no one has gotten around to analyzing…. so during the pandemic, we started digging into those data and developing visualization modules and we realized we can actually make this into an app so that we can feed more data and generate visualizations
The team behind eVITTA
To discuss the different aspects of eVITTA and to delve deeper into this project, UBC’s Shayan Abbaszadeh sat down for a virtual interview with PhD candidate Judith Yan from the department of Cell and Developmental Biology at UBC. Dr. Yan is one of the many faces behind eVITTA at the Taubert lab and has worked tirelessly to bring this idea into fruition. Our podcast describes her role in realizing the gaps in efficient transcriptome analysis and building eVITTA with the rest of the Taubert Lab during the COVID-19 pandemic. Outside of eVITTA, Dr. Yan’s lab work in the Taubert Lab usually involves using model organisms such as roundworms to study stress responses and applying that knowledge to better understand human diseases.
How does eVITTA make analyzing RNA so simple?
To grasp the scope of this research, first and foremost, it is important to understand transcriptome analysis. Dr. Yan describes transcriptome analysis as a powerful tool that examines how RNA, a copy of DNA, is used in a cell, tissue or organism. This involves taking out the whole RNA from a sample, getting it sequenced (i.e. decoded), and subsequently obtaining information from that data.
An exploration of gene patterns is one of the main aspects involved in effective transcriptome analysis. A gene is a section of DNA that carries a specific piece of information. A crucial aspect of analysis is understanding that some genes are turned on at higher rates than others. According to Dr. Yan, once there is a count for these different genes, the numbers can be interpreted to reveal useful information about “what some of these genes are doing” and “what processes and gene sets are actually being changed“. Finally, there needs to be effective visualization techniques of the different data sets and the data needs to be validated against previously published data.
All of these functions are achieved through eVITTA; a user-friendly, web-based interface that streamlines the multiple steps of transcriptome profiling. Watch this short video to become familiar with the 3 modules of eVIITA; easy-GEO, easyGSEA, and easyVizR, and realize what effective transcriptome analysis looks like!
The challenges of transcriptome analysis and how evitta addresses them
In our podcast, we focused extensively on the motivations behind the creation of eVITTA, specifically in relation to challenges associated with transcriptome analysis and how eVITTA deals with these challenges in ways that previous methods could not. Dr. Yan alludes to the use of the ‘overrepresentation approach’ (ORA) in previous technologies that has the flaw of only being able to represent a small subset of gene changes. As Dr. Yan puts it: “you’re missing out on a lot of information and biologically… you are not able to capture the less severe changes“. eVITTA on the other hand, focuses on entire sets of genes instead of just one gene, allowing scientists to observe gene changes across the board.
Additionally, previous technologies did not allow for the organization of data because “oftentimes it is very tedious to do multiple comparison because you have many different subsections that you want to look at“. eVITTA prevents the dumping and mislabeling of gene data which expedites the process of discovering important biological patterns. This platform has already been proven to be highly effective in studies involving severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and roundworms (C.elegans).
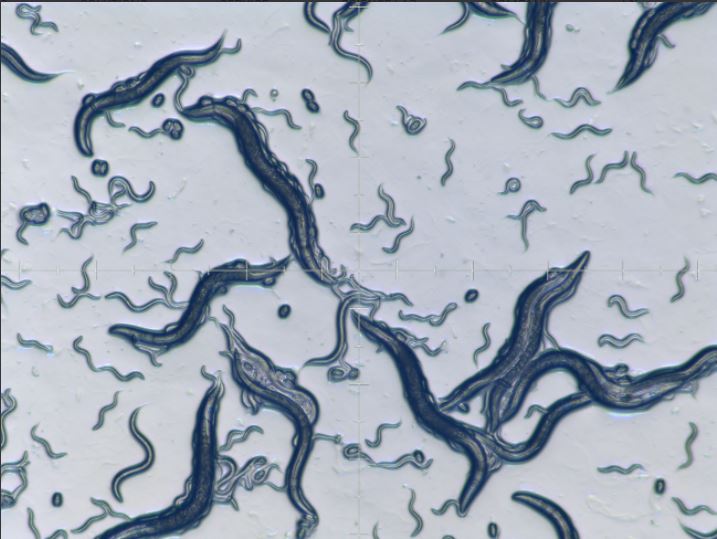
Dr. Yan’s research using eVITTA involves the starvation response of C.elegans roundworms. (credit: Taubert lab)
what does the future hold for eVITTA?
Our podcast also discusses the future of eVITTA and the team’s plans for further expansion. Dr. Yan and her team plan to add more algorithms to analyze the expression of different genes at a more in-depth level. The team also want to add more visualization options to provide more choice for gene dataset analysis. Finally, they are planning to update eVITTA’s databases by adding more datasets to the application. These ideas and more are discussed in our podcast below!
As transcriptome profiling provides major insight regarding diseases and illnesses, the optimization eVITTA can provide may be increasingly vital for today’s society. For instance, if transcriptome analysis can be conducted more efficiently, crucial findings in disease mechanisms may be discovered sooner. As a result, treatments and health regulations can be created more quickly, potentially saving lives and preventing disease spread. Additionally, eVITTA’s user-friendly and web-based interface makes transcriptome profiling accessible to more biologists around the world and would therefore greatly benefit the research community.
– Heather Cathcart, Kaushali Ghosh, Parham Asli, Shayan Abbaszadeh



 from lets Talk science
from lets Talk science from Walmart
from Walmart


 https://d2r55xnwy6nx47.cloudfront.net/uploads/2021/02/Brain-Noise_2560_Lede.jpg
https://d2r55xnwy6nx47.cloudfront.net/uploads/2021/02/Brain-Noise_2560_Lede.jpg https://i.guim.co.uk/img/media/e16e33bdce44e28c845484cf9b93efafbdcac570/0_171_5121_3073/master/5121.jpg?width=620&quality=45&auto=format&fit=max&dpr=2&s=1b96aff2315ed8ee9176c6908c50f57b
https://i.guim.co.uk/img/media/e16e33bdce44e28c845484cf9b93efafbdcac570/0_171_5121_3073/master/5121.jpg?width=620&quality=45&auto=format&fit=max&dpr=2&s=1b96aff2315ed8ee9176c6908c50f57b https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4380153/figure/fig-1/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4380153/figure/fig-1/