Have you ever experienced a problem that R failed in recognizing part of your time series data, and analysed them by omitting thousands of rows in your spreadsheet?
You then looked for the cause, and found that the time format of your data seemed to have subtle inconsistency in Excel. Some parts of the time data were Text-type (e.g., from row 335 to 344 in Fig. 1), while the other parts were Datetime-type (e.g., from row 324 to 334 in Fig. 1), though these data were downloaded from the same data logger at the same time.
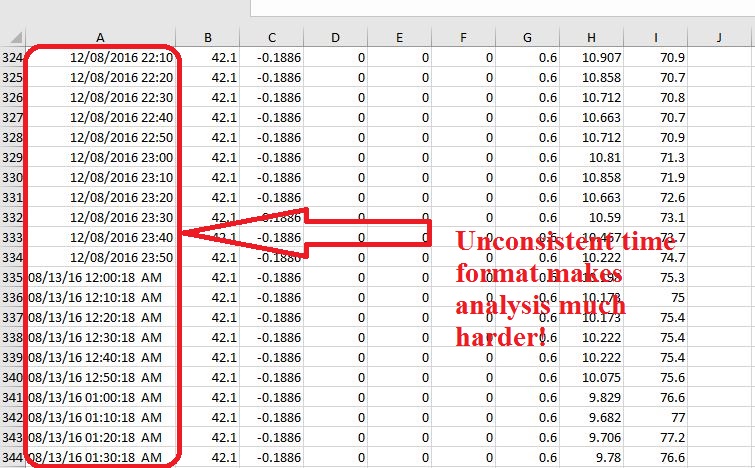
Fig. 1 A screenshot of my data, illustrating the inconsistency time format problem.
So you tried to make the time format consistent, by selecting the whole column (e.g., Column A in Fig. 1), right-clicking, entering the Format Cells box, and choosing the desired format (e.g, in my case, I chose custom format, mm/dd/yyyy h:mm).
However, you found that this didn’t work on your Text-type data. So you tried again, and unfortunately, failed again. You even tried different types of paste and copy, but had no effect. After tens of failures, you gave up and decided to manually re-type time data row by row into the spreadsheet to amend the format, but “millions” of Text-type data which embellished in your good Datetime-type data here and there drove you crazy. You started to worry that you would be stuck in typing data for the rest of your life.
What’s worse, the Datetime-type data were also problematic, because Excel recognized months as days, (e.g., in Row 334 in Fig. 1, 12 is actually the day). You could also not be able to correct them by just changing the format setting. What a pissed-off moment!
Fig. 2 Only Rage Face can express my "rage" on Excel. Hahahaha....
But don’t be frustrated! I had fought with this moment many times, and eventually defeated it. Now, I’d love to share my tips with you on how to deal with this problem.
Why did this problem happened?
I have searched for the cause for a long time, and found that people who claimed the similar problems are most in Canada. So I speculated that the problem came from system regional setting. Later, I found other peoples’ explanations on the causation, which confirmed my speculation.
According to their explanations, if you imported US format (mm/dd/yyyy) time series data from your data logger into your computer which set as Canadian English/French format (dd/mm/yyyy), then Excel would mistake days as months when the days are under 12, while leave the rest (the days are greater than 12) as unrecognized, text-type data.
Here are two discussions I found on Stack overflow (http://stackoverflow.com/questions/4660906/some-dates-recognized-as-dates-some-dates-not-recognized-why)
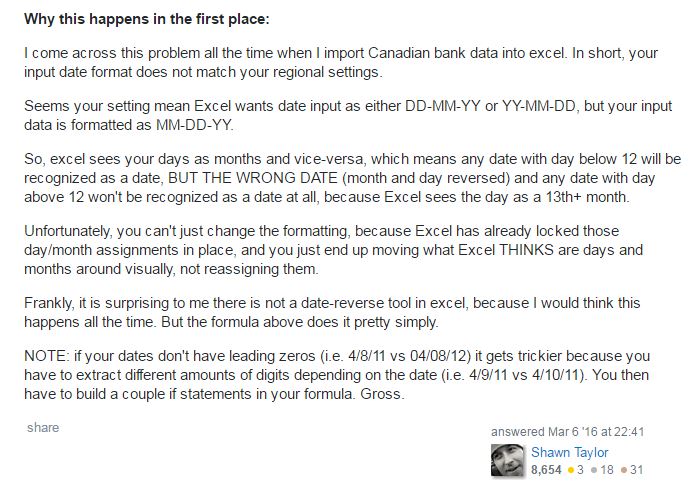
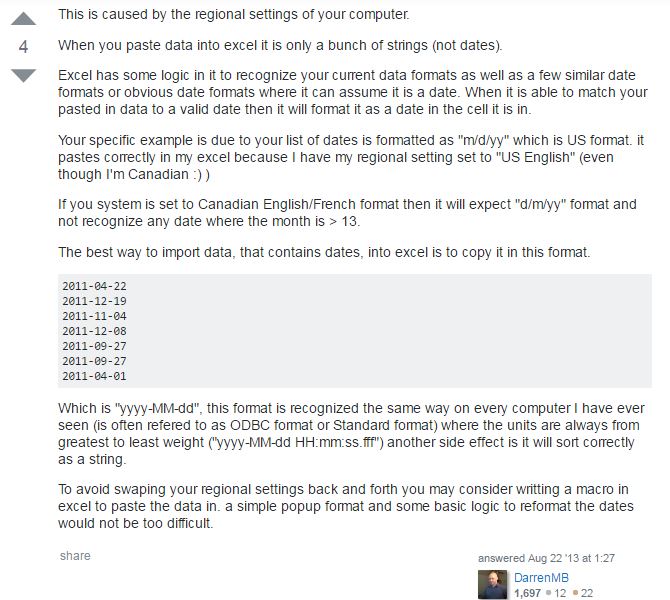
Fig. 3 Other peoples' discussion on the same issue at Stack Overflow.
How to deal with this problem?
Firstly, check date settings in your computer.
Click the time display at the right bottom of desktop to enter the Date and Time box, and click Change date and time…, entering Date and Time setting, and then click Change Calendar Setting, entering Region and Language box. Under Format tab, you can modify time format under Short date and Long date. In my case, I prefer MM/dd/yyyy to dd/MM/yyyy as the short date format in my computer.
This step helps you to avoid any further problem, if you need to import your data from external sources such as your data logger.
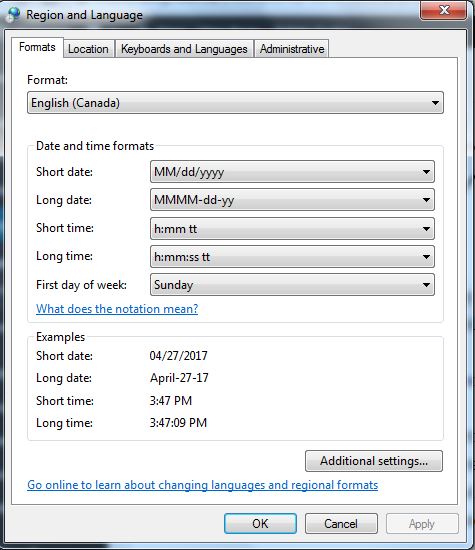
Fig. 4 Time format setting in the Window system.
Secondly, modify the wrong Datetime-type data.
(1) Change your wrong Datetime-type data into Text type data, by using TEXT Function. For example, the data in Column A, Row 329 is 12/08/2016 11:00:00 PM, with a custom format of mm/dd/yyyy. I use function: =TEXT(A329, “dd/mm/yyyy H:MM”) to convert it into Text-type data. Please note that I use format “dd/mm/yyyy” instead of “mm/dd/yyyy”, in order to exchange “months” with “days”.
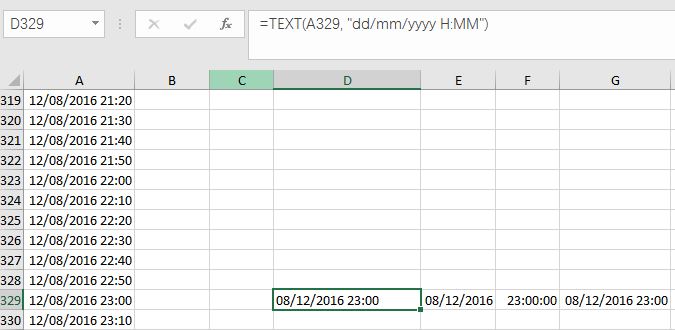
Fig. 5 My illustration on how to convert Datetime-type data into Text-type data.
(2) Convert Text-type data into Date-type and Time-type data,by using DATE Function and TIME function. I used =DATE(MID(D329,7,4), LEFT(D329,2), MID(D329,4,2)) and =TIME(MID(D329,12,2),MID(A337,14,2),0) in Cell (E, 329) and Cell (F, 329) separately. Please note that in my case, I don’t need seconds, so I put 0 in the time function. Then I gathered Date and Time together at Cell (G, 329) by simple addition: E329+F329. You can also complete those steps at one time by putting DATE and TIME functions together in one cell.
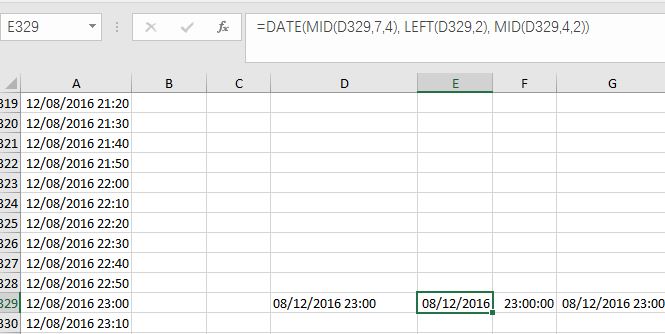
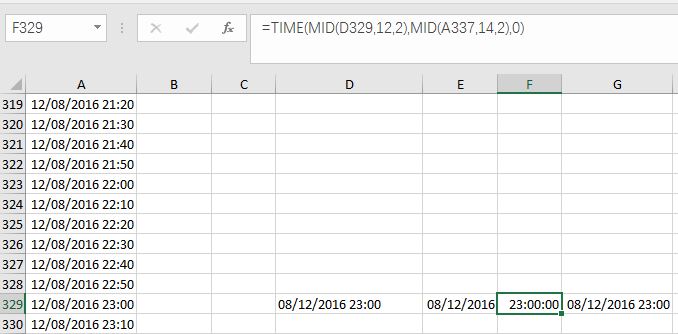
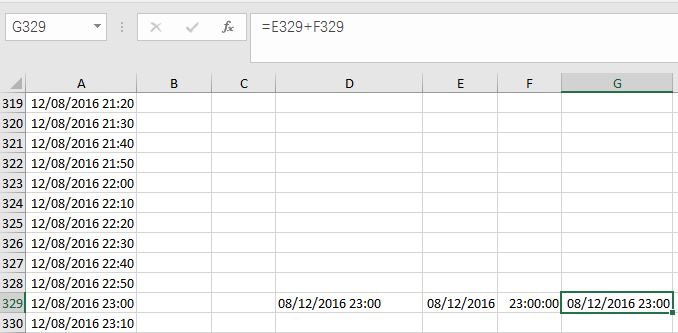
Fig. 6 My illustration on how to convert Text-type data into Datetime-type data.
Here is a detailed introduction on TEXT, DATE and TIME Functions:
Format Example
TEXT (Cell, “desired format”) ==> TEXT(A329, “dd/mm/yyyy H:MM”)
DATE (year, month, day) ==> DATE(MID(D329,7,4), LEFT(D329,2),MID(D329,4,2))
TIME (hour, minute, second) ==> TIME(MID(D329,12,2),MID(A337,14,2),0)
In addition, here are the how the extract functions like MID and LEFT work:
MID (Cell, The location of first character to extract, the number of extracted characters)
MID Function extracts characters from inside of the string. For example, MID(D329,7,4) is equal to “2016”, because the “2” of “2016” in Cell D329 is the 7th characters, and 2016 consist of four characters.
LEFT (Cell, the number of extracted characters)
LEFT Function extracts characters starting from the left of the string. For example, LEFT(D329, 2) is equal to “08”, because “08” are first two characters at the left.
Please note that space or symbols like “/”, “:” or one space “”, are counted as one character.
Thirdly, convert other Text-type data into Datetime-type data by using the same functions introduced above.
You may encounter a new problem when your Text-type data contains “am” or “pm”. Unfortunately, I didn’t find a function to extract “am” or “pm” into 24-hour system. But you can use subtraction to overcome this problem.
For instance, Cell D337 is “08/13/16 12:10:18 AM”. If I use TIME(MID(A337,10,2),MID(A337,13,2),MID(A337,16,2)), then Excel gives me the result as “12:10:18” in the 24-hour system, which is “12:10:18 pm”.
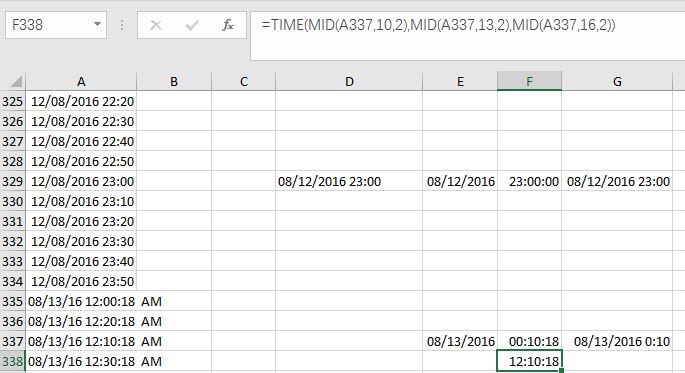
Fig. 7 Illustration on how to deal with "am" or "pm" in your data.
So instead, I used TIME(MID(A337,10,2) -12,MID(A337,13,2),MID(A337,16,2)), and Excel gave me the exact result that I want, the “00:10:18”.
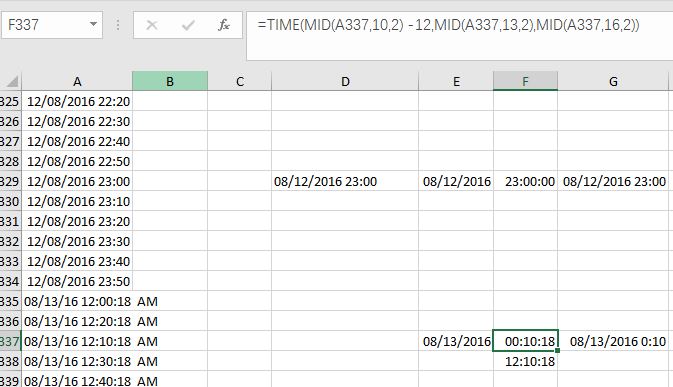
Fig. 8 Illustration on correct your time data by simple subtraction.
With above three steps, you can successfully arrange your “naughty” time series data in the “stupid” Excel. Life turns beautiful again!

If you have know more convenient ways to deal with this problem, please tell me by leaving your comments here. 🙂
Thank you for reading it!
April, 28, 2017
Fig.1 Main road that was still covered by snow
Fig. 2 The intermittent creek at B1T1 (left) and at the path along B1C (right)
Fig. 3 B1C
Fig. 4 B1T2
Fig. 5 Climate station at B1T1


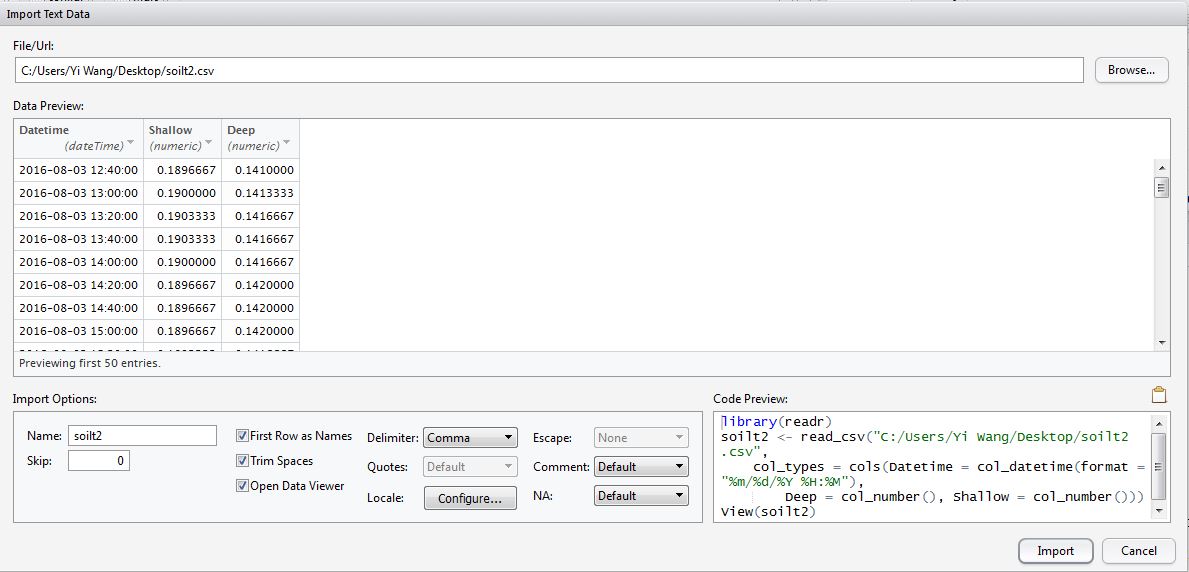
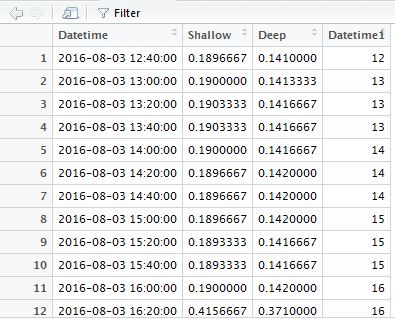
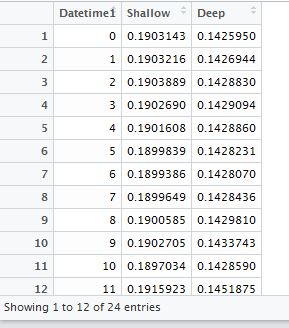
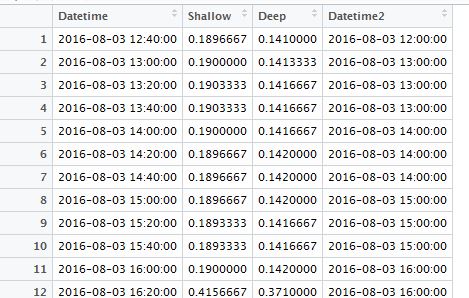
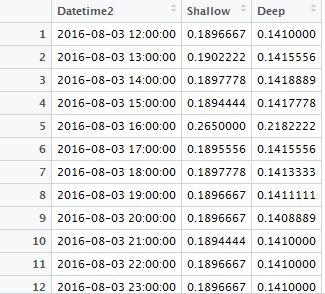












 Fig. 1 The road to my experiment site was covered by snow
Fig. 1 The road to my experiment site was covered by snow Fig. 2 Photo of B1T1 took at Oct, 18, 2016
Fig. 2 Photo of B1T1 took at Oct, 18, 2016 Fig. 3 the band dendrometer sensors were taken back, leaving metal clamp and spring on the site.
Fig. 3 the band dendrometer sensors were taken back, leaving metal clamp and spring on the site. Fig. 4 the datalogger were taken back, and the wires were protected by plastic films
Fig. 4 the datalogger were taken back, and the wires were protected by plastic films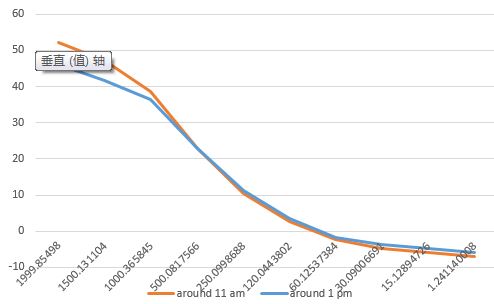


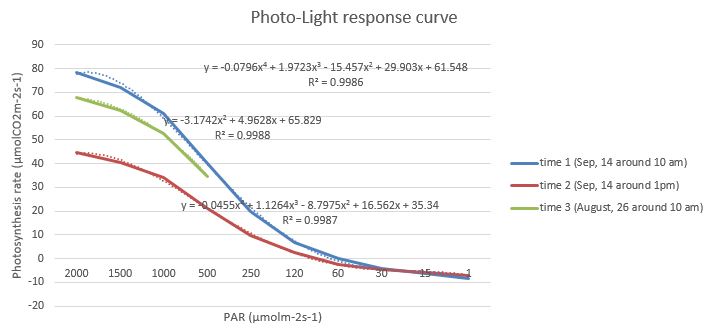 Fig. 1 Light-responsive curves of photosynthesis rate
Fig. 1 Light-responsive curves of photosynthesis rate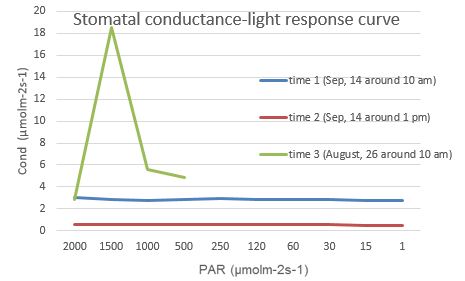 Fig. 2 Light-responsive curves of stomatal conductance
Fig. 2 Light-responsive curves of stomatal conductance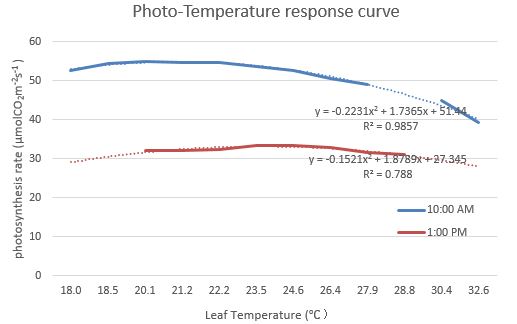 Fig. 3 Temperature response curves of photosynthesis rate
Fig. 3 Temperature response curves of photosynthesis rate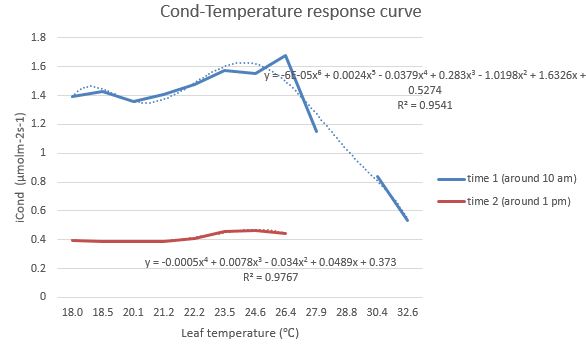 Fig. 4 Temperature response curves of stomatal conductance
Fig. 4 Temperature response curves of stomatal conductance
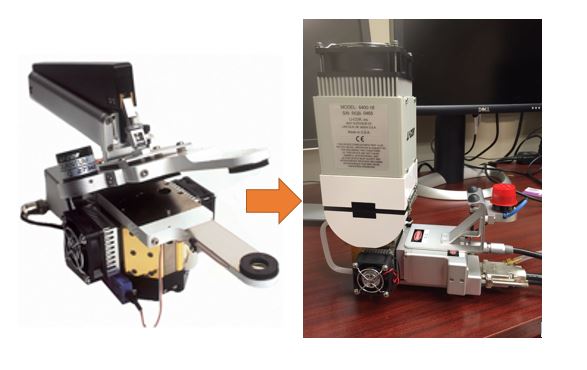 Fig. 1 I assembled opaque connifer chamber
Fig. 1 I assembled opaque connifer chamber