Since the onset of genetic studies, scientists have noticed that classical Mendelian mechanisms alone could not explain the phenotypic patterns observed in living organisms. Waddington coined the word epigenetics in 1942 while studying how gene expression patterns are modified during differentiation and development. Here is the mental picture he had in mind:
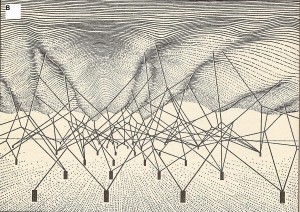
The upper surface is the phenotypic landscape. The anchored pegs symbolize genes, and strings pulling the landscape are genetic interactions that shape the phenotypic landscape.
(Source: openi.nlm.nih.gov)
Waddington’s successors brought great improvement in the understanding of epigenetic mechanisms during the last century. Epigenetics can be seen as an optional turning on and off of genes, a specific regulation pathway that plays a crucial role in plant and animal development. Gene expressions can be regulated through changes in the condensation level of the chromatin or methylation of promoters. Methylation seems to be the main process involved in plant epigenetics, and involves the interaction between RNA polymerases, small RNAs and DNA-binding proteins (see Henderson et al. 2007 for more precise explanations and clear illustrations of the molecular mechanisms).
In parallel to developmental studies, epigenetics has been extensively studied by evolutionary biologists as a mechanism of transgenerational non-genetic determination of phenotypes. New epigenetic marks can be triggered by environmental conditions. They are subsequently either transmitted to the next generation or lost during reproduction. The study of these features of epigenetic inheritance unveiled striking examples of plastic adaptive responses to unstable environments, like the development of protective helmets in Daphnia in the presence of predators (Agrawal et al. 1999).
Have you ever thought that a microscopic animal could be cooler than X-men?
(Source: discovermagazine.com)
As epigenetic mode of inheritance seems to be a widespread and valuable strategy to fine-tune adaptive traits in fluctuating environments, it is reasonable to suggest that such mechanisms could be involved in adaptation to climate in tree populations, especially widespread and high-dispersal species. And indeed, it is the case in Norway spruce (Picea abies).

A Norway spruce cone. An epigenetic mechanism in the seed partly determines phenology traits of the future tree.
(Source: www.flickriver.com)
Scientists from the Norwegian Forest and Landscape Institute have been studying the phenomenon for 30 years (Yakovlev et al. 2012), ever since managers of Norwegian forests reported that individuals from the central part of the range grafted in southern orchards did not perform as expected. They found that the temperature at some stage during sexual reproduction was the main environmental trigger influencing seedling performance, in addition to day length. Further studies suggest that the epigenetic mark is not transmitted through parental gametes but acquired during seed formation in response to day length and temperature. There is therefore an epigenetic memory of climatic conditions in the embryo during its development in the mother tree. This memory influences the subsequent seedling’s phenology and performance. This series of studies on Norway spruce shows that epigenetic processes can be an important factor influencing adaptation to climate in tree populations. Learning more about the relative importance of epigenetic and genetic processes in adaptation studies will certainly help predicting the adaptive potential of natural populations facing climate change, as well as improving conservation and forest management plans.
There is still a lot to discover about epigenetic processes, how they relate to and interact with genetic processes, and what influences the maintenance or loss of epigenetic marks among generations. Along with the exponential development of genomic studies thanks to technical improvements, epigenomic studies now allow looking at variations in DNA condensation levels and methylation patterns at the scale of whole genomes. This is what Schmitz et al. (2013) did in a recently published study. They looked at epigenomic variation patterns in different populations of Arabidopsis thaliana and determined how they relate to genetic variation at the scale of the genome. Even though the results don’t give a clear view of how often epigenetic states are associated with phenotypic variation, such a study improves our understanding of how association studies that include the epigenome can help to uncover additional sources of heritability and contribute to adaptation processes.
References
Agrawal, A. A., Laforsch, C., & Tollrian, R. (1999). Transgenerational induction of defences in animals and plants. Nature, 401(September).
Henderson, I. R., & Jacobsen, S. E. (2007). Epigenetic inheritance in plants. Nature, 447(7143), 418–24. doi:10.1038/nature05917
Schmitz, R. J., Schultz, M. D., Urich, M. a., Nery, J. R., Pelizzola, M., Libiger, O., Alix, A., et al. (2013). Patterns of population epigenomic diversity. Nature, 495, 193–198. doi:10.1038/nature11968
Yakovlev, I., Fossdal, C. G., Skrøppa, T., Olsen, J. E., Jahren, A. H., & Johnsen, Ø. (2012). An adaptive epigenetic memory in conifers with important implications for seed production. Seed Science Research, 22(02), 63–76. doi:10.1017/S0960258511000535

very interesting blog. I´ll be looking fwd to more interesting articles in the future