The amount of plastic that has been produced to date now exceeds 8300 million metric tonnes (Mt). To put this into perspective, the average blue whale weighs approximately 180 Mt, thus 46 million blue whale’s worth of plastic has been produced since humans started commercializing plastics in 1950. Our society had become extremely dependent on plastic products and synthetic (petroleum- based) textiles which cause serious consequences such as microplastic and microfiber pollution as I’ve discussed in previous blog posts.

Figure 1. Size reference for blue whales. Wikipedia Commons
Bioplastics have more recently taken the stage as a potential avenue for replacing petroleum-based plastics. Biologically based polymers have structural elements such as helices, β-turns, β-sheets and coils which provide structural integrity and resilience and can replicate the desirable polymeric interactions in plastics. Additionally, a lot of new material is being developed based on the protein polymers that are naturally occurring in biological systems (including ourselves).
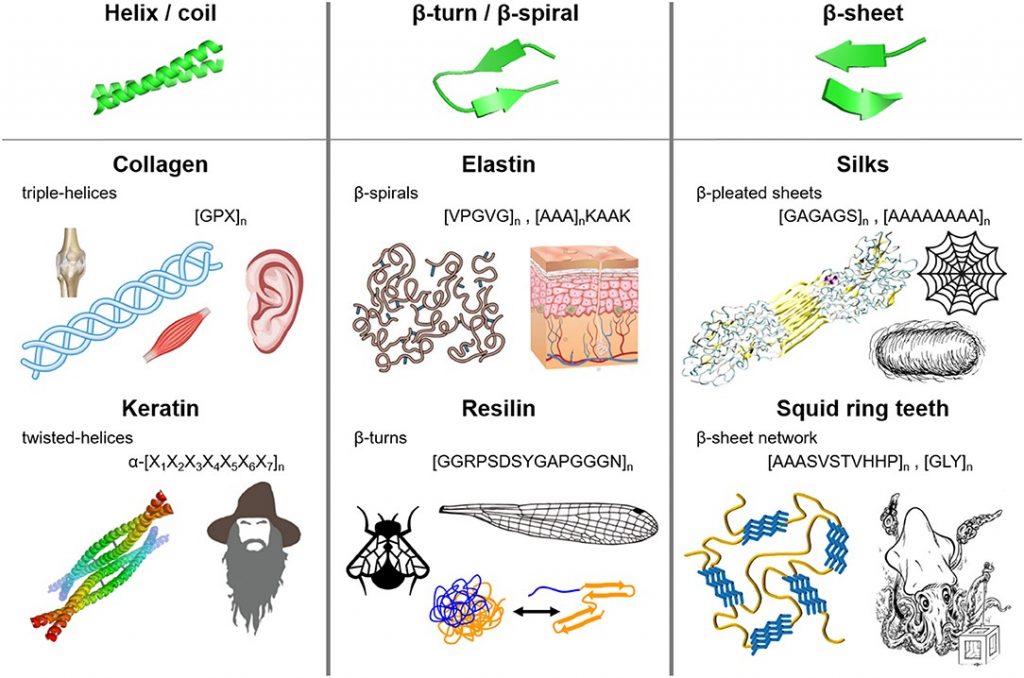
Figure 2. Fibrous protein polymers have molecular architecture that can include (i) helices and coils, (ii) β-turns and β-spirals, and (iii) β-sheets. Source
Video 1. SRT- coated fabrics that self-heal in water.
Squid ring teeth (SRT) are an especially promising candidate for making functional fibres and films due to its strength, conductivity and self-healing properties. The SRT are located inside the suction cups of the tentacles of squids and are composed of a naturally occurring protein complex. Fortunately, it is not necessary to harvest squid to obtain the SRT proteins as they can be biosynthetically produced since having their genome sequenced. The SRT-inspired monomer is repeated to create a polymeric chain and the resulting protein that is produced is named accordingly as tandem repeat (TR) proteins.
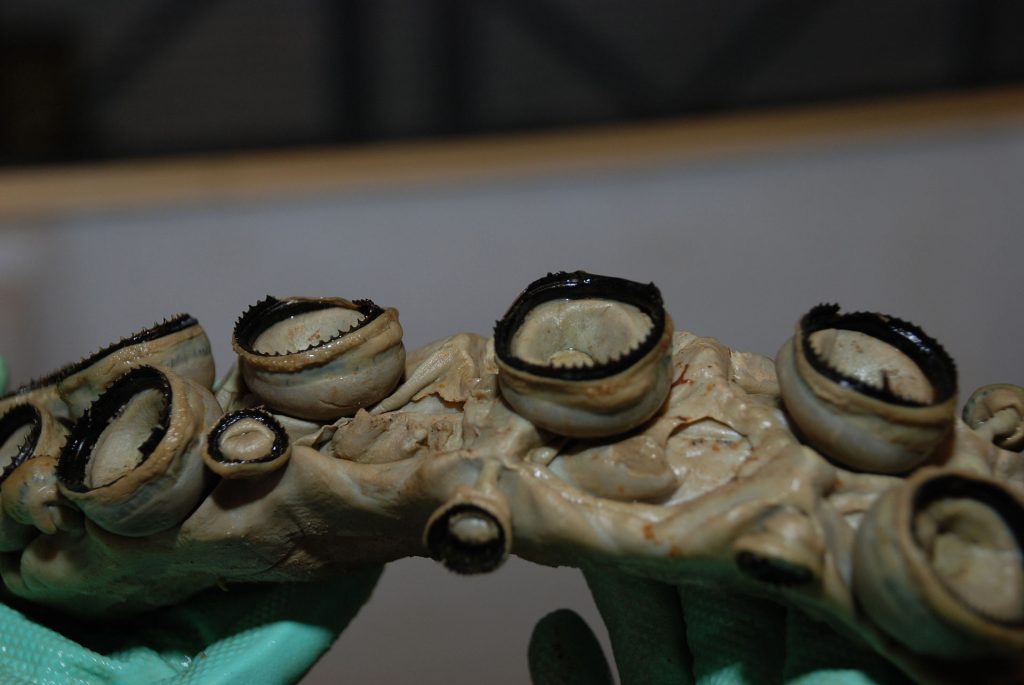
Figure 3. The Squid ring teeth of a giant squid. Wikipedia Commons
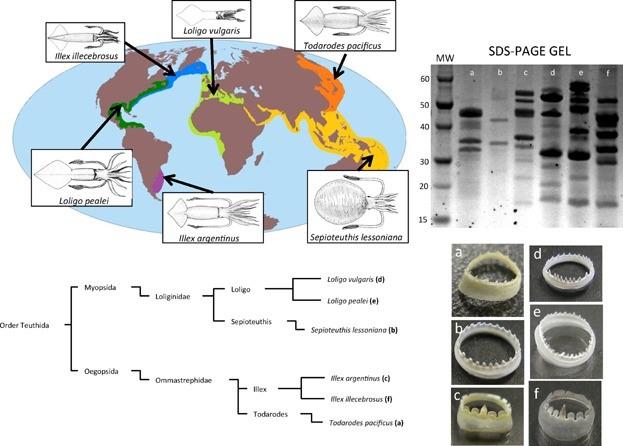
Figure 4. Optical images of squid ring teeth (SRT) and the six common squid species they originate from. Source
Films produced with SRT proteins consist of disordered domains that provide elasticity and flexibility to the materials in addition to ordered domains such as β-sheets that provide mechanical strength. One study designed four TR proteins denoted as TR-nX where X was the number of repeat units within the molecule. They measured the mechanical force of these four TR proteins and found that the ultimate strength of the protein’s scale linearly, with TR-n25 reaching an ultimate strength of 40 MPa. As seen in Figure 4 below, there is a limitation of the study due to sample size. The error bars of the TR-n12 and TR-n25 overlap, so it is not possible to say there is a statistically significant difference from each other.
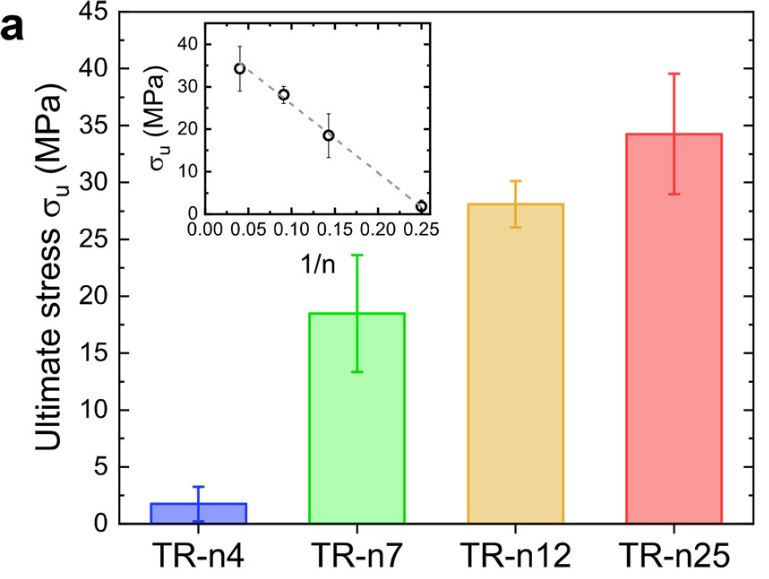
Figure 4. Mechanical testing of fully hydrated TR proteins (inset shows 1/n dependence) Source
Additionally, SRT protein films have been suggested as a potential solution to the issues related to the release of microfibers into the water from washing. A cloth made from polyester was coated with an SRT protein film and was found to dramatically increase the cloths resistance to abrasion (and microfiber release) compared to cloths that were not coated with SRT protein films.
While there is still a lot of further research to be done, SRT-based proteins are a promising avenue for making our world a little bit less plastic.
~Isla