Introduction
Asthma is one of the most prevalent diseases among children and affects more than 300 million people worldwide. The respiratory illness is difficult to diagnose since there is no set pattern for symptoms and those symptoms can vary widely depending on each patient and their environment. Many people who suffer from asthma develop the chronic disease when they are young and often need to adhere to steadfast treatment throughout their life. In addition, asthma can’t be completely prevented as the exact cause is still unknown.

Title: Living With and Managing Asthma
Courtesy of the U.S. National Institute of Health via Wikimedia Commons
New Hope
However, scientists at the University of British Columbia have published new research that is raising hopes of easing asthma’s grip on the world. The work was spearheaded by Dr. Marie-Claire Arrieta and Leah Stiemsma from the Department of Microbiology and Immunology. In a world’s first, their team discovered evidence of a potential relationship between the risk of childhood asthma and early microbial changes in one’s gut.

Published in Science Translational Medicine (September 2015)
Our Science Outreach Group (Luxi Xu, Lisa Kim, Paul Yi, King Biong) sought out Dr. Arrieta and Stiemsma to discuss their research and the implications it has for future diagnostics and therapies for asthma patients. Check out our video below for a general overview of the study and what it means for the general public.

Extended Interview Clips with Dr. Marie-Claire Arrieta

Experimental Design & Methods: Eureka!
The Vancouver-based team attained their pioneering results by studying human subjects and conducting animal experiments. Fecal samples from a total of 319 infants were analyzed for their gut microbiota (the ecological community of microorganisms that share our lower digestive tract). The young subjects were classified into three categories for the research study.
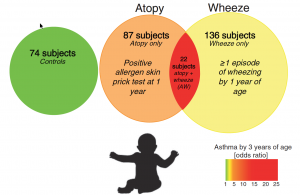
Fig. 1 Classification of study participants
Source: Arrieta 2015 via Science Translational Medicine
The data showed that there was a transient gut microbial imbalance for the infants who were at risk of asthma during the first 100 days of their lives. Infants at risk of asthma presented a significant decrease in the abundance of four specific bacterial genera: Lachnospira, Veillonella, Faecalibacterium, and Rothia (also referred to as “FLVR” as a group).
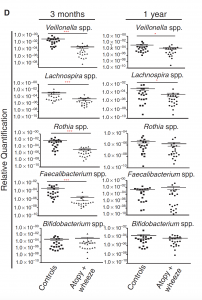
Fig. 2 (D) Gut microbial composition by clinical phenotype at 3 months and 1 year (infants)
Source: Arrieta 2015 via Science Translational Medicine
To study the role of these four bacterial taxa, researchers injected the microbes in question into germ-free mice (mice that are bred to have no bacteria in their gut). Surprisingly, the bacterial taxa appeared to help by relieving lung inflammation in the lab mice compared to the control group.
Baby Steps Toward Progress

The infant subjects were selected from the Canadian Healthy Infant Longitudinal Development (CHILD) Study. Partly funded by the Canadian Institutes of Health Research, this unique, public health study spans across the country from British Columbia to Ontario. More than 3,500 families are signed up to assess their children from birth until they reach five-years-old. The aim is to create a comprehensive database of genetic and environmental triggers in order to understand the root causes of allergies and diseases.

Media Attention
The results have attracted considerable attention in the media from the Vancouver Sun to CBC News. Even drawing a positive outlook from TIME, The Washington Post, and The Wall Street Journal. It was interesting to compare how each news outlet reported on the story and what implications the research held. For example, the Wall Street Journal introduced its story with the threat of the “explosive” rise of asthma over the past 50 years. The Vancouver Sun, however, opened with the hope of a potential cure. It also gave a chance to hear from other senior researchers who oversaw the study.
“What this study helps emphasize — and there’s a growing awareness of this recently — is that we need to rethink our relationship with bacteria …”
– Dr. Stuart Turvey, Director of Clinical Research at CFRI.
Study Limitations
Like all research, this study has faced a number of limitations. Stiemsma pointed out that the study wasn’t designed to account for confounding variables such as breastfeeding, cesarean births, or antibiotic use. Rather, the conclusion was there is now evidence that supports the hypothesis that changes in the gut microbiota affect the risk of asthma development. Whether these influential factor, such as antibiotics, play a direct role in the onset of asthma has yet to be determined.
Working with children also poised its own challenges. Collecting blood samples from a 3-month-old infant would not be feasible; biological samples for each subject were not always available, raising the level of variability of when the samples were taken. In contrast, the mice experiment had a high level of control.
Hypothetically, a more efficient way to determine whether certain bacteria affect the development of asthma would be to directly inoculate children with the FLVR microbes. Unfortunately, such a procedure would be unethical. Dr. Arrieta echoed this sentiment, describing it as too simplistic and unsafe to “bombard” babies with an unknown range of bacteria.
“So we really need to figure out what the bacteria are doing and more importantly how much of the are needed because with the microbiota, it’s not as simple as just adding bacteria….” – Dr. Claire Arrieta
Going Forward
Dr. Arrieta emphasized that further research should be focused on studying the mechanisms of how the four bacterial taxa work which may confirm whether they can be used for a potential cure for asthma or a substantial treatment. Stiemsma added that their results will be need to be validated at a much larger scale, potentially including all 3,000 infants in the CHILD Study.
From Parents’ Perspective:
Check out our podcast for further information on prevention of asthma development in young children from a parent’s perspective: