Hello team, please see below for my revised definition of polymerase chain reaction (PCR). PDF file of the definition can be found here: 301 Janice Pang Technical Definitions_revised.
Introduction
The purpose of this assignment is to learn how to write definitions for a specific audience that may not have the technical knowledge of the term being defined. This assignment serves to highlight the importance of definitions in technical writing and how audience and purpose indicate the need for definitions.
The Situation
In the fall, Janice will be starting her new role as a biology teacher at a high school. For the biology curriculum at her school, she must introduce a common laboratory technique used in molecular biology to senior students with limited biology background. She has chosen to teach her class about polymerase chain reaction. As such, she needs to prepare a definition for polymerase chain reaction that will be handed out to students in class and posted on the class website for reference.
Parenthetical Definition
Polymerase chain reaction (PCR; reaction used to make many copies of DNA) is commonly used in molecular biology experiments.
Sentence Definition
Polymerase chain reaction (PCR) is a molecular biology technique used to generate millions or billions of copies of a specific segment of DNA.
Expanded Definition
Polymerase Chain Reaction (PCR)
What is PCR?
PCR is a common molecular biology technique used to amplify (make many copies of) specific regions of DNA in the test tube (Karp, 2013). PCR provides researchers a way to discover functions of genes (Karp, 2013). The aim of PCR is usually to make enough of a certain target DNA region to support subsequent analysis using other methods (Reece et al., 2010). For example, the DNA can be sent for sequencing or cloned in bacteria for further experimentation (Karp, 2013). By using PCR, many copies of specific segments of DNA can be made within a few hours, faster than any methods previously developed (Reece el al., 2010).
What is the origin of PCR?
The origins of PCR traces back to the early 1980s at Cetus Corporation in California, where many of the key research for the development of the technology was performed (Bartlett & Stirling, 2003). At the time, researchers wanted to find a simple method to generate large amounts of DNA (Mullis, 1990). In 1983, while driving in a Honda Civic on Highway 128 from San Francisco to Mendocino, Kary Mullis conceived of the idea to develop a new method to amplify specific segments of DNA (Mullis, 1990). This technique became known as PCR (Mullis, 1990). The emergence of PCR transformed fields in the life sciences. PCR formed the foundation of the human genome project and became fundamental in numerous molecular biology laboratory protocols (Bartlett & Stirling, 2003).
What is needed for PCR?
PCR requires elevated temperature, DNA template, all four DNA nucleotides, DNA polymerase, and primers. This technique only occurs in higher temperatures as the two strands of DNA must separate before copying can begin (Reece et al., 2010). The DNA template used in the reaction contains the specific DNA sequence being copied (Reece et al., 2010). The DNA template is used by DNA polymerase, which creates new strands of DNA (Reece et al., 2010). The DNA polymerase typically used in PCR is the heat stable Taq polymerase found in bacteria species,Thermus aquaticus, from hot springs (Chien, Edgar, & Trela, 1976). DNA polymerase is able to create new strands of DNA using the four nucleotides, adenine, thymine, guanine and cytosine, that make up the DNA double helix (Karp, 2013). For specificity, PCR needs primers, which are short pieces of single-stranded DNA at least 15 nucleotides long (Reece et al., 2010). There are two primers used per PCR reaction, each pairing to the ends of the opposite strands of the DNA template (Reece et al., 2010). Collectively, in elevated temperature, primers bind to the DNA template, DNA polymerase extends the primers and a specific region of DNA is copied.
How does PCR work?
The key steps of PCR are shown in Figure 1. For PCR to occur, the key ingredients (DNA template, primers, four DNA nucleotides and DNA polymerase) of the reaction are mixed in a test tube along with other cofactors required by the enzymes and put through cycles of heating and cooling that enable DNA to be synthesized (Karp, 2013). The basic steps of PCR are:
- Denaturation (95°C) – sample is heated briefly to separate DNA into two strands.
- Annealing (60°C) – reaction is cooled to allow primers to pair to the target region of the DNA template.
- Elongation (72°C) – temperature is raised to allow DNA polymerase to add nucleotides to the end of the DNA template, generating new strands of DNA.
These three steps are repeated continuously for about 30 more cycles, each time doubling the amount of DNA (Karp, 2013). Consequently, billions of copies of a specific segment of DNA are created.
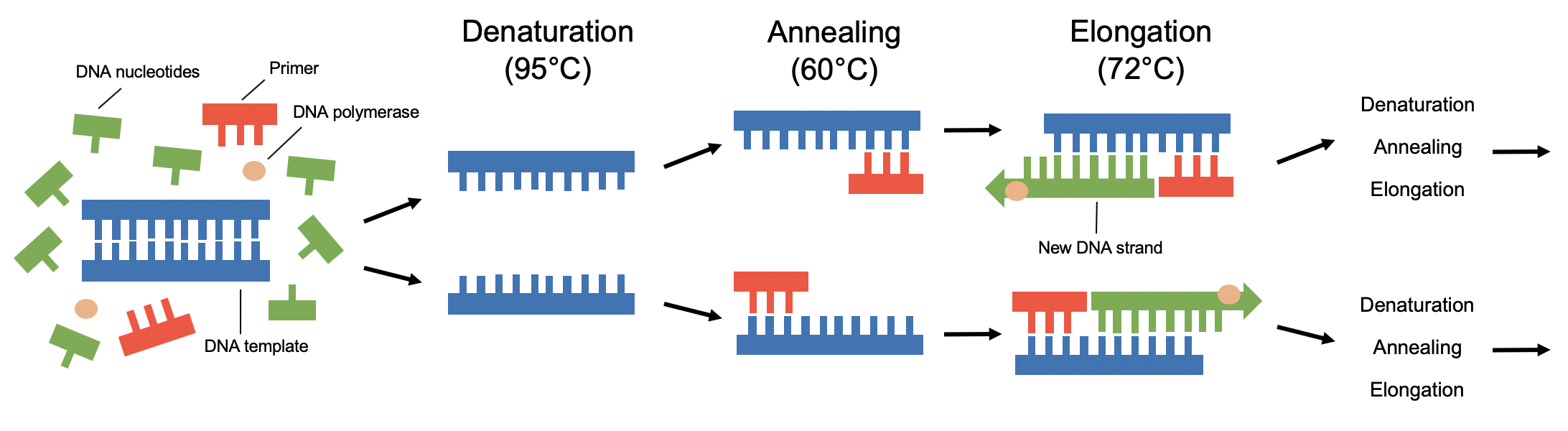
| Figure 1 Polymerase chain reaction (PCR). Key steps of this reaction are denaturation, annealing and elongation. These three steps are repeated for around 30 cycles to generate large amounts of DNA. |
What are the applications of PCR?
PCR has a wide range of applications. For example, this technique can support genomic studies whereby researchers can send the DNA generated from PCR for sequencing (Garibyan & Avashia, 2013). PCR can also be useful for quantifying or testing for presence of specific DNA in biomedical research aimed at understanding gene functions (Karp, 2013). PCR has even led to the development of medical diagnostic tools that allow for accurate and rapid detection of potentially life-threatening infectious diseases caused by bacteria and viruses (Yang & Rothman, 2004). Furthermore, PCR can assist forensic science, where small amounts of DNA can be isolated from a crime scene, copied and compared to DNA databases to identify criminals (van Pelt-Verkuil, van Belkum, & Hays, 2008).
References
Bartlett, J. M. S., Stirling, D. (2003). PCR Protocols (2nded.). Totowa, New Jersey: Humana Press Inc.
Chien, A., Edgar, D. B., & Trela, J. M. (1976). Deoxyribonucleic Acid Polymerase from the Extreme Thermophile Thermus
aquaticus. Journal of Bacteriology, 127(3), 1550–1557.
Garibyan, L., & Avashia, N. (2013). Polymerase Chain Reaction. Journal of Investigative Dermatology, 133(3), 1–4.
Karp, G. (2013). Cell and molecular biology concepts and experiments (7thed.). Hoboken, New Jersey: John Wiley & Sons, Inc..
Mullis, K. B. (1990). The Unusual Origin of the Polymerase Chain Reaction. Scientific American, 262(4), 56–65.
Reece, J. B., Urry, L. A., Cain, M. L., Wasserman, S. A., Minorsky, P. V., & Jackson, R. B. (2011). Campbell Biology (9thed.). San
Francisco, California: Pearson Benjamin Cummings.
van Pelt-Verkuil, E., van Belkum, A., & Hays, J. P. (2008). Principles and Technical Aspects of PCR Amplification. Springer.
Yang, S., & Rothman, R. E. (2004). PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in
acute-care settings. The Lancet Infectious Diseases, 4(6), 337–348.
Leave a Reply