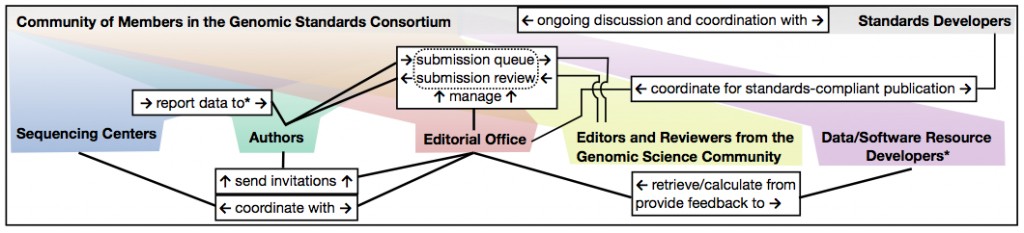
An Open-Access, Standards-Supportive Publication that Rapidly Disseminates Concise Genome and Metagenome Reports in Compliance with MIGS/MIMS Standards: the Session Blog
Presenters: Oranmiyan W. Nelson
July 9, 2009 at 2:30 pm
Background
Dr. Nelson is currently completing his post-doctoral work at Michigan State University, in the Department of Microbiology & Molecular Genetics. He is also the production editor of Standards in Genomic Sciences (SIGS), an open-access e-journal of the Genomics Standards Consortium (GCS) established in September 2005.
Session Overview
Genomics and metagenomics describes the study of an organism’s complete genetic data set, with emphasis on sequence mapping. The former is limited to the study of organisms that can be isolated in pure culture, whereas the latter applies to an entire community of microbes in their native environment. Technological advancements have facilitated a data explosion; genomic maps are now being produced at a faster rate than the existing publication infrastructure can accommodate. The result is a loss of data, contextual metadata and annotation. Essentially valuable data is disappearing before it can be interpreted or see the light of publication.
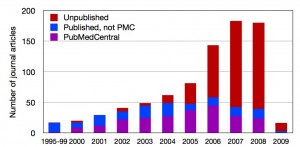
Number of published journal articles per year relating to the complete genome sequences of bacteria and archea
SIGS is a GSC initiative to attempt to bridge this gap, by producing concise peer-reviewed reports that comply with MIGS/MIMS standards, in addition to operation procedures, commentary and review articles. Minimum Information about a (Meta)Genome Sequence (MIGS/MIMS) is a GCS initiative to expand on core reporting standards already established by the International Nucleotide Sequence Database Collaboration (INSDC). MIGS/MIMS is characterized by a standardized checklist that has been published in its entirety in Nature Biotechnology. SIGS will attempt to keep up with data production by offering an accelerated editorial revision workflow, where copy-editing begins almost immediately after initial acceptance.
The e-journal’s goal is to produce 600 publications by June 2011 (approximately 30 per month), by attracting internationally credible authors under a cost-effective model for an open-access journal.
A sample short genomic report was then described, in conjuction with MIGS/MIMS standards. The anatomy of a report included a abstract and introduction (a), genetic sequencing information (b) genome properties (c) and comparisons with previously sequenced genomes (d). Lastly, conclusions and references (e).
References