Many people may have heard about cutting and pasting genes from a genome, the complete blueprint of an organism, into microorganisms to make them magically produce compounds that they could never produce naturally. However, few experiments have endeavored to make a novel organelle, an artificial factory within a living cell. Genetic evidence shows that the mitochondrion, the cell’s power plant, was actually a bacterium that got swallowed by an ancestral cell, and it revolutionized how life works. Wouldn’t it be extremely cool if scientists could do something like that and change how cells behave?
Organelles are compartments performing specific functions, found within the cells of many organisms. They concentrate reactants for chemical reactions while not disturbing the cell’s other activities. For example, the organelle called peroxisome is a processing plant and protector of plant and animal cells. It degrades very long chain fatty acids from dietary fats and produces toxic hydrogen peroxide, which is safely reduced to water within. Zellweger syndrome is due to dysfunctional peroxisomes, where a person accumulates of toxic metabolites and cannot make myelin for nerve cells and bile for fat digestion, which may be fatal with no available treatment. Therefore, it is important to maintain healthy peroxisomes and their protein machineries.
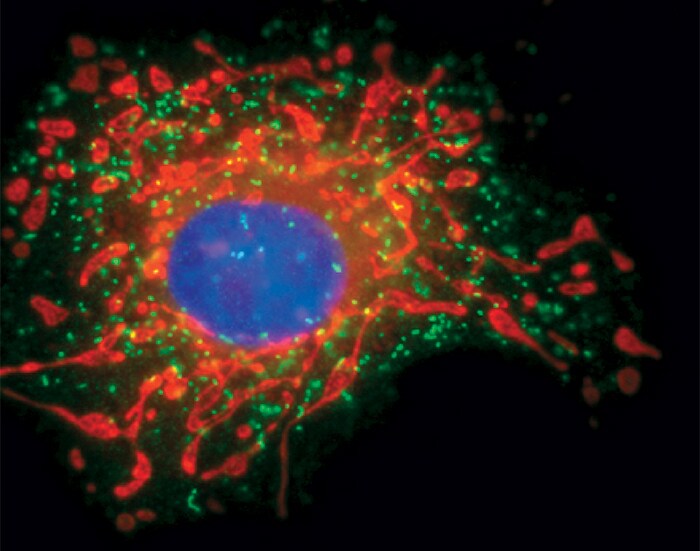
Structure of Peroxisome in fixed cells, labelled with SelectFX® Alexa Fluor® 488 Peroxisome Labeling Kit. Adapted from Thermo Fisher Scientific: Peroxisome Structure
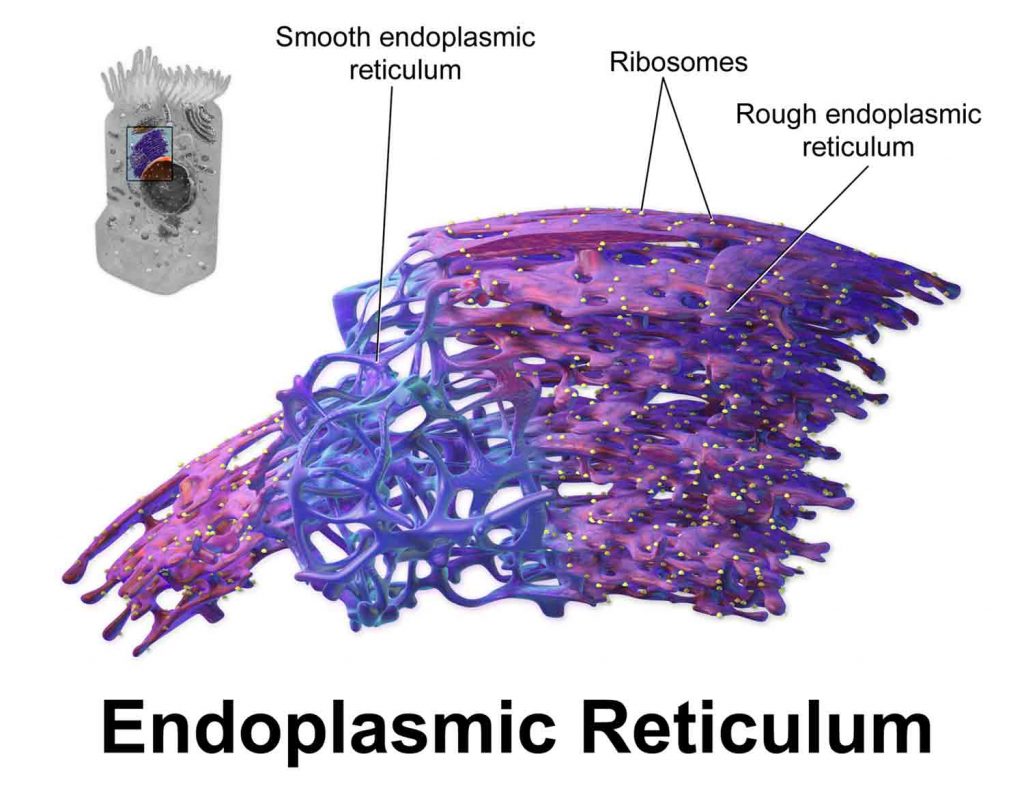
Interestingly, peroxisome proteins are imported directly from the cytosol, the fluid-filled space surrounding organelles within the cell, without going through a central protein-sorting system in another organelle, the web-like endoplasmic reticulum.
In September 2017, chemical biologist Stuart Warriner and his team at the University of Leeds, England, published their work on tweaking the natural peroxisomal protein import system by modifying two proteins involved in the pathway. They found that their new pathway overrides the natural protein carrying pathway with no adverse side effects for the cell. Theoretically, scientists could design it in whichever way they want through this simple, subtle change.
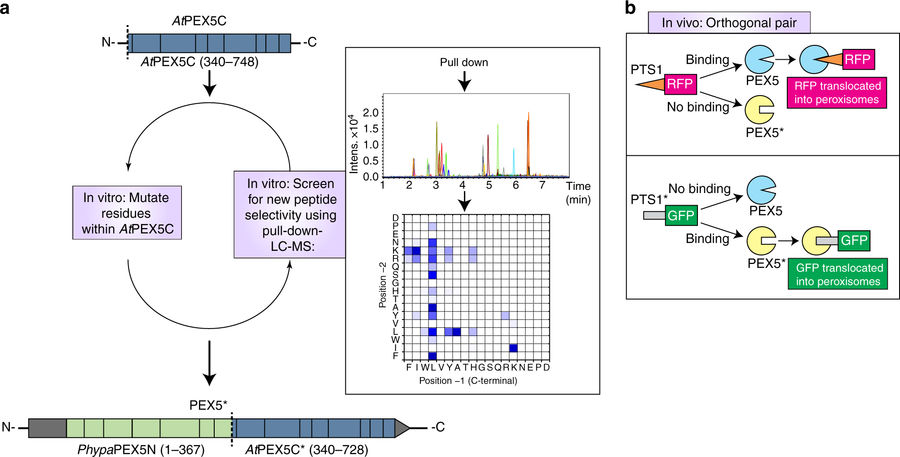
Figure 1. Scheme of research. Adapted from Figure 1 of “Towards designer organelles by subverting the peroxisomal import pathway” by Warriner et. al, 2017.
A big family of proteins called PEX (peroxins) serve as porters and receptors that recognize and dock cytosolic proteins with the five-letter amino acid peroxisomal targeting sequence (PTS; like a password) to import them into peroxisomes. However, the researchers only changed the PTS and a protein called PEX5, which wanders in the cytosol looking for homeless, PTS-carrying proteins.
Using known crystal structures of PEX5 protein, the researchers generated several versions of PEX5 by mutating (changing) one or two amino acids at one end that binds the original PTS. They modified the PTS by changing its last two amino acids, and identified a new PTS that rcognized the new PEX5’ but not the original.
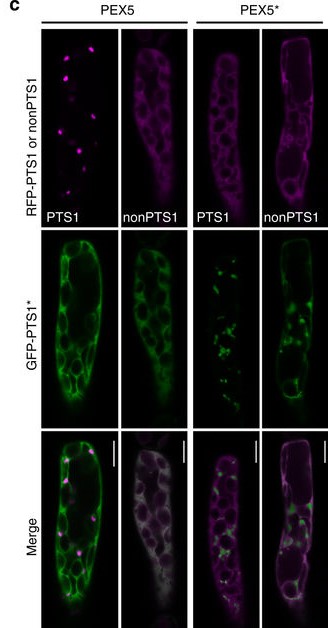
Adapted from Figure 3 of “Towards designer organelles by subverting the peroxisomal import pathway” by Warriner et al., 2017, showing how the new PEX5 protein selectively imports new PTS-containing proteins into peroxisomes.
Unexpectedly, viewing cells with fluorescence microscope showed the natural PTS was imported by the new porter protein PEX5’, meaning PEX5’ is more lenient on PTS. However, when fluorescent PTS and PTS’ were inserted in PEX5′-containing cells, only the new PTS’ ended up in peroxisomes, leaving the PTS in the cytosol. Evidently, the new and original PTS compete for import, with the original PTS outcompeted – how they interact with other PEX proteins and by what mechanism are still mysteries. However, this means scientists can manipulate peroxisomes by simply switching to the new PEX5′ protein.
Custom organelles has many potential applications, such as more efficient and sustainable production of therapeutics and other synthetic compounds without compromising the cell’s other activities and health. Scientists could also design a novel waste management system for the cell by designing PTS-containing proteins that scavenges certain chemical wastes from the cytosol and import them for processing in peroxisomes. Without knowing when that day will come, meanwhile, we should appreciate what our wonderful organelles do to keep us alive!
Written by Jenny Zhong