Essential Idea: Genetic information in DNA can be accurately copied and can be translated to make the proteins needed by the cell.
DNA structure and replication crash course
Transcription and Translation crash course
Protein Synthesis slides (Bio 115)
DNA Replication – Basics
2.7.U1 -The replication of DNA is semi-conservative and depends on complementary base pairing.
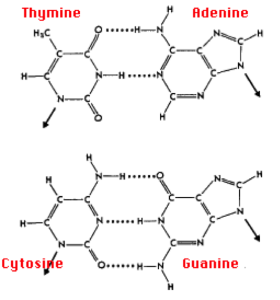
Two DNA strands are held together by hydrogen bonding between complementary base pairs.
- Adenine is always matched with Thymine with two hydrogen bonds
- Guanine is always matched with Cytosine with three hydrogen bond
- This is due to the molecular structure of the base pairs.
- Base paring ensures two identical DNA strands are formed after replication is complete
- The parental strand acts as a template and therefore each replicated strand is identical and each daughter cell has the same DNA.
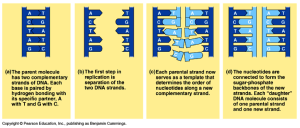
Due to this, DNA replication is semi-conservative – or each new DNA strand has one parental strand (template) and one newly synthesized strand.
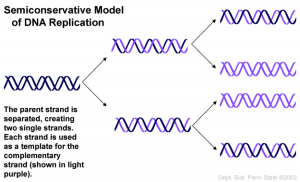
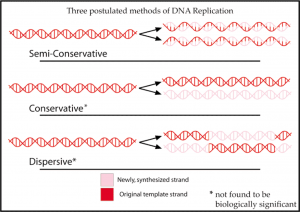
How do we know DNA replication is semi-conservative?
- Originally, there were three proposed methods:
- 1) Conservative – parental strand rejoins after replication
- 2) Dispersive – parental DNA double helix is broken into segments that act as templates
- 3) Semi-conservative – two parental DNA strands separate and each of those strands then serves as a template for the synthesis of a new DNA strand.
In 1957, Meselson and Stahl conducted an experiment providing evidence for semi-conservative replication.
2.7.A2 – Analysis of Meselson and Stahl’s results to obtain support for the theory of semi-conservative replication of DNA.
Meselson and Stahl experiment:
- DNA is semi-conservative in terms of replication.
- Using the parental DNA strand as a template, two identical strands of DNA are produced.
- But, in order for base pairing to occur, we need to unzip to DNA.
DNA Replication
DNA molecule is split down the middle and forms two, identical copies
2.7.U2 – Helicase unwinds the double helix and separates the two strands by breaking hydrogen bonds.
Steps of Transcription:
1. Initiation
- DNA Helicase (an unzipping enzyme) moves along DNA molecule breaking hydrogen bonds between complementary base pairs.
- Replication bubble – Unwound/open region of a DNA helix where replication occurs.
- Separated DNA strands form a replication bubble at the origin of replication
- Initiator protein binds to an origin of replication and triggers unzipping (several ori
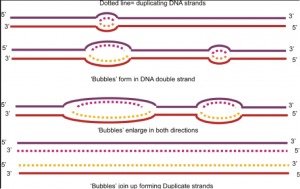
Figure 6: DNA replication bubble gins of replication)
- As helicase unzips the DNA, strain on the helix increases.
- Must relieves tension periodically.
- Topoisomerase – Enzyme that cuts DNA and reseals it with fewer twists (relieves tension).
- We now have two template (parental) strands with bases exposed. Complementary base paring can occur.
2.7.U3 – DNA polymerase links nucleotides together to form a new strand, using the pre-existing strand as a template.
2. Elongation
- Free-floating nucleotides form complementary base pairs with the expo
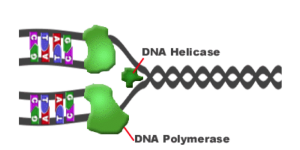
Figure 7: DNA replication enzymes sed bases of the unzipped parental strand.
- DNA Polymerase (enzyme) helps covalently bond the new paired bases together (sugars and phosphate groups of nucleotides)
- DNA Polymerase always works in a 5’ to 3’ direction
Question: If DNA polymerase always works in a 5’ to 3’ direction, what does these mean for the newly synthesized strands?
Each strand is synthesized in opposite directions of each other – One will run continuously -> moves in the same direction as helicase unzips. The other will have to be synthesized discontinuously -> moves in the opposite direction as helicase unzips. To elaborate:
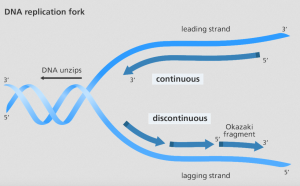
Leading strand – 3’ to 5’
- Nucleotides can readily be added to the newly synthesized strand in the opposite 5’ to 3’ direction.
- Continuous synthesis (it occurs in the direction of DNA being unzipped i.e. replication fork)
Lagging strand – 5’ to 3’
- The nucleotides are added in the opposite direction of the DNA unzip (i.e. away from the replication fork)
- Discontinuous synthesis (It is “lagging” behind)
Question: If the enzyme moves away from the fork, and the fork is uncovering new DNA that needs to be replicated, then how can the lagging strand be replicated at all?
- Okazaki fragments are short segments of DNA synthesized in the 5’ to 3’ direction (away from the unzip/replication fork)
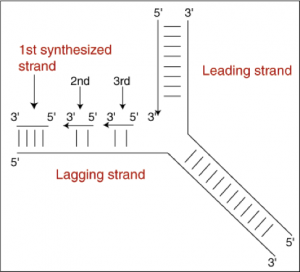
Figure 9: Lagging strand replication - Each subsequent piece is replicated more closely to the replication fork than the fragment before it.
- DNA Ligase (another enzyme) joins all the Okazaki fragments together
Protein synthesis and the Central Dogma
Central dogma – Explains the flow of genetic information within a biological system.
- “DNA makes RNA and RNA makes protein”
- Two processes: Transcription and Translation
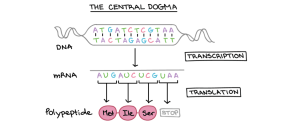
Transcription
Transcription is the process by which an RNA sequence is produced from a DNA template.
- Sections of DNA that code for a specific polypeptide are called genes.
- Gene = specific DNA sequence at specific location
- The DNA code is in the nucleus, but protein synthesis takes place outside nucleus on ribosomes.
Question: What needs to occur before protein synthesis can take place?
- There must be an intermediary molecule that carries DNA code outside of the nucleus.
- mRNA (Messenger RNA) is this molecule!
2.7.U4 – Transcription is the synthesis of mRNA copied from the DNA base sequences by RNA polymerase.
Steps of transcription:
RNA Polymerase – An enzyme that unzips a specific area of DNA (gene).
- Iniation
- RNA polymerase binds to specific sequence called
- RNA Polymerase begins to unzip an area of DNA.
- Elongation
- RNA nucleotides found in the nucleus are added to the template strand of the DNA by RNA polymerase (C-G, A-U) in the 5’ to 3’ direction. This is called elongation.
- RNA polymerase covalently bonds nucelotides of mRNA strand.
Things to consider (elongation):
- RNA is single stranded, therefore only one strand of DNA is used as a template.
- DNA strand used as template = the template or antisense (3′ to 5′) It is complementary to mRNA.
- Unused DNA strand = the coding or sense strand (5′ to 3′) and has the same sequence as the mRNA.

- Termination
- RNA polymerase reaches terminator (specific sequence that signals end of transcription) and mRNA strand detaches.
- Processing
- Eukaryotic genes are composed of protein-coding sequences called exons (exons are expressed) and introns (intervening)
- Intron sequences do not encode functional proteins.
- Therefore, introns must be removed by a complex molecular machine called a spliceosome – this process is called
- mRNA leaves the nucleus via the nuclear pore.
2.7.U7 – Codons of three bases on mRNA correspond to one amino acid in a polypeptide.
The genetic code is written in triplets (3 base code)
DNA —–> Transcription —–> mRNA
Triplet Codon
Each codon codes for one specific amino acid.
Question: If one codon (3 bases) code for 1 amino acid, what must the entire mRNA strand code for?
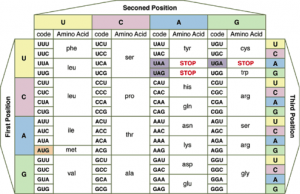
Translation
Translation is the process of protein synthesis in which the genetic information encoded in mRNA is translated into a sequence of amino acids in a polypeptide chain.
2.7.U5 –Translation is the synthesis of polypeptides on ribosomes.
2.7.U6 – The amino acid sequence of polypeptides is determined by mRNA according to the genetic code.
Translation takes place at the ribosomes (in cytoplasm or on the rough ER)
Messenger RNA (mRNA) carries information from a specific gene to the ribosomes in order to create the correct polypeptide.
Types of RNA:
mRNA (messenger RNA) single-stranded complementary copy of template DNA strand coding for a single polypeptide.
rRNA (ribosomal RNA) non-coding RNA which makes up ~60% of a ribosome composition. Helps decode mRNA into amino acids through interactions between mRNA and tRNA.
tRNA (transfer RNA) carries one of the 20 amino acids to the ribosome for polypeptide formation.
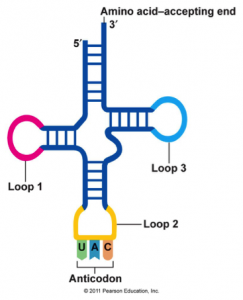
- Contains a sequence called the anticodon, which can recognize and decode a specific complementary mRNA codon.
- Each tRNA has its corresponding amino acid attached to its end.
- When a tRNA recognizes and binds to its corresponding codon in the ribosome, the tRNA transfers the appropriate amino acid to the end of the growing polypeptide chain.
- Resembles a clover.
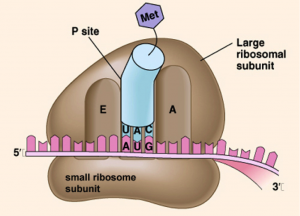
Ribosome structure:
- The ribosomes consist of a large and a small subunit.
- Each ribosomal subunit is made of rRNA and protein.
- The small subunit binds to mRNA.
- The large subunit has binding sites for tRNA. Also, catalyzes peptide bonds between amino acids.
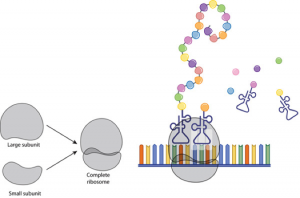
Steps of Translation:
1.Initiation
- The small ribosomal subunit binds to the mRNA strand.
- The mRNA strand is “sandwiched” between the small and large ribosome subunits.so that the first two codon triplets are within the boundaries of the ribosome.
- Complementary tRNA molecule joins with the first codon of the mRNA molecule (i.e. must have an anti-codon complementary to the mRNA codon) (Which subunit contains tRNA binding sites?)
- The first codon, or start codon, is AUG which codes for Methionine (Met). (Peptidyl (P) site on the large ribosomal subunit)
Question: What would be the tRNA anti-codon for Methionine?
2. Elongation
- The next complementary tRNA anti-codon matches with the mRNA codon in the Aminoacyl (A) site.
- The two amino acids carried by the tRNA are side by side now.
- A condensation reaction occurs , covalently bonding the P site amino acid to the A site amino acid.
- The P site tRNA is released, the ribosome slides down the mRNA (A site tRNA is now at the P site), and a complementary tRNA anti-codon matches with the next mRNA codon at the now empty A site.
- The ribosome only advancemence one codon at a time.
- To summarize – The ribosome travels down the mRNA, reading codons and bringing in the complementary tRNA’s to translate the message out to protein.
3. Termination
- Elongation continues until the ribosome reaches a STOP codon.
- The ribosome accepts a protein called a release factor instead of tRNA at the A Site.
- The release factor breaks the bond between tRNA and the polypeptide – releasing both.
Central Dogma summary,
- DNA -> RNA -> PROTEIN
- DNA is transcribed into single stranded mRNA molecules with the help of the enzyme RNA Polymerase.
- The single stranded mRNA is complementary to the template (3’ to 5’) strand and identical to the coding (5’ to 3’) strand (Except replace T with U on mRNA)
- The synthesized mRNA molecule is able to leave to nucleus via the nuclear pore and heads to the ribosomes – the site of protein synthesis and translation.
- The two ribosomal subunits bind to the mRNA strand.
- tRNA carry specific anti-codons which are complementary to the mRNA codons. The tRNA binds mRNA codon, carrying a specific amino acid.
- The ribosome moves along the mRNA strand adding amino acids (translating the mRNA into a polypeptide).
- When a stop codon is reached, a release factor binds, releasing the polypeptide.
- Polypeptides are part, or the whole, of a specific protein which can have many functions around the body.
Comments by shaun pletsch