Essential idea: Every living organism inherits a blueprint for life from its parents.
3.1.U1 – A gene is a heritable factor that consists of a length of DNA and influences a specific characteristic.
Introduction to genes
Gene = Heritable factor that consists of a length of DNA and influences a specific characteristic.
Heredity = the passing on of characteristics genetically from one generation to another.
Remember: DNA has four base pairs Adenine, Thymine, Guanine, Cytosine. The specific sequence of the base pairs codes for a specific polypeptide, or protein. Proteins play many critical roles around the body but are required for the structure, function, regulation of body tissues and organs.
3.1.U2 – A gene occupies a specific position on a chromosome
- Humans have ~ 21,000-23,000 protein coding genes
- Each gene is at a specific location.
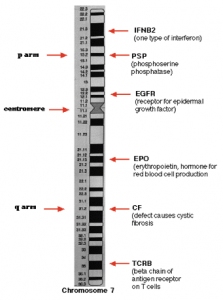
- To elaborate, Each gene occupies a specific location or position on a chromosome. This position is called a locus (plural loci)
Humans have 46 chromosomes
- 22 autosomal pairs and 1 pair of sex chromosomes (23 pairs = 46)
- ~ 21,000-23,000 protein coding genes packed into 23 pairs of chromosomes
- How do humans stack up?
3.1.A2 – Comparison of the number of genes in humans with other species.
3.2.A2 –Comparison of genome size in T2 phage, E.coli, D. melanogaster, H. sapiens and Parascaris equorum
|
Organism (Common name) |
Organism (Scientific name) |
Genome size (# of bases) |
Number of genes |
|
Virus (bacteriophage) |
T2 Phage |
5400 |
11 |
|
Bacterium |
Escherichia coli |
4 639 000 |
4377 |
|
Nematode (roundworm) |
Caenorhabditis elegans |
100 292 000 |
20 000 |
|
Asian rice |
Oryza sativa |
430 000 000 |
Up to 56 000 |
|
Fruit fly |
Drosophila melanogaster |
122 654 000 |
27 407 |
|
Human |
Homo sapiens |
3 000 000 000 |
~21 000 |
|
Japanese canopy planet |
Paris japonica |
150 000 000 000 |
? |
NOTE: The number of genes in a species should not be referred to as genome size as this term is used for the total amount of DNA.
3.1.U3 – The various specific forms of a gene are alleles.
Differences in genes
- Humans share ~99.9% of DNA
- However, in every given human there are approximately 3 million differences in individual bases
- These differences create specific forms of genes, or (i.e. variations or versions of genes)
- There can be two or more alleles of a specific gene, depending on the gene.
- Remember that humans have two copies of each gene – one maternal and paternal.
3.1.U4 – Alleles differ from each other by one or only a few bases
- These alleles only have to differ by one or a few base pairs to completely alter the characteristic.
- For example,
| Where the gene is found | Sequence of bases | Protein and trait |
| Locus for transducin gene on chromosome 1. | …CACCAGCAACCC… | Transducin protein synthesized, colour vision possible |
| …CACCAGTAACCC… | Protein not formed correctly, colour vision not possible |
- Two alleles for the transducin protein are present in humans – a single base difference causes the transducin protein to not be formed correctly in some individuals and therefore colour vision is not possible. (Complete achromatopsia)
- This means that a single trait (ability to distinguish colour) can have variants (i.e. alleles!)

Evolution of phototransduction in vertebrates [website – complex]
Another example we will be looking at in the future is blood type. There are three alleles for blood types: A, B, and O.
3.1.U5 – New alleles are formed by mutation. [Deletions, insertions and frame shift mutations do not need to be included]
Mutations
- A mutation is a random, rare change in genetic material.
- This can occur during replication or do to environmental factors.
- Mutations can give rise to new alleles of a particular gene.
- As you learned, alleles only have to differ by one or a few bases to completely alter the characteristic.
- Mutations that cause a change in a single nucleotide base are called point mutations. (Point mutation is a random SNP – Single-nucleotide polymorphism)
Point mutations can have four affects:
- Silent Mutation – Mutation that does not change the amino acid sequence of a protein.
- Missense Mutation – Mutation that results in a codon that codes for a different amino acid.
- Nonsense Mutation – Mutation that results in a premature stop codon. The protein incomplete – it is nonsense! Analogy: Think about reading Harry Potter – everything is going great but suddenly the book ends with him on the train to Hogwarts. Does the movie make sense? Or is it nonsense?
- Read through Mutation – Mutation that results in a stop codon being changed into a different amino acid. (The mRNA sequence can be read through – past the stopping point.)
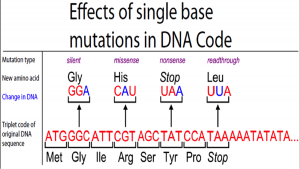
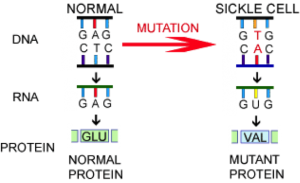
-This single base substitution (right) changes Thymine to Adenine on the antisense DNA strand. This in turn alters the mRNA codon, which alters the amino acid sequence (Glutamic acid -> Valine), which changes the composition and structure of the resulting protein.
– A single base mutation (point mutation) resonates throughout the central dogma and affects gene expression.
This particular point mutation creates an allele that affects the hemoglobin in red blood cells.
- Individuals who have this allele have sickle cell disease.
Question: What type of point mutation leads to production of sickle cell hemoglobin?
3.1.A1 – The causes of sickle cell anemia, including a base substitution mutation, a change to the base sequence of mRNA transcribed from it and a change to the sequence of a polypeptide in hemoglobin. [Students should be able to recall one specific base substitution that causes glutamic acid to be substituted by valine as the sixth amino acid in the hemoglobin polypeptide.]
Sickle Cell Disease
- A single base mutation changes the mRNA codon for glutamic acid into one that codes for valine (GAG ->GTG) at the sixth position of the sequence.
- This alters the composition and shape of the resulting protein, creating a sickle shaped blood cell.
Symptoms:
- Sickle shaped red blood cells cannot carry oxygen as efficiently and therefore weakness, fatigue, and shortness of breathe are all symptoms of sickle cell disease.
- Hemoglobin tends to crystalize in sickle cells, causing them to become more rigid and get stuck in capillaries, blocking the flow of blood. Affected individuals may also suffer strokes, blindness, or damage to the lungs, kidneys, or heart. They must often be hospitalized for blood transfusions.
- Many suffers will die before the age of 20.
Question: Sickle cell disease typically leads to premature death among suffers, however it is still prevalent (~1 in 100 people in Africa). Why has such a fatal disease been able to persist at a relatively high frequency?
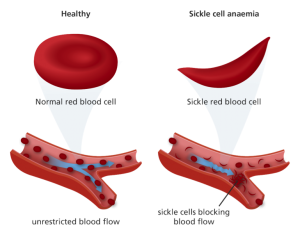
Advantages:
- Although debilitating, individuals with sickle cell disease are resistant to malarial infection.
- Malaria is caused by a parasite, Plasmodium, that is transmitted to humans by the Anopheles The parasite infects and grow on red blood cells, eventually causing the cell to burst. Symptoms include high fever, chills, and potentially death.
- Recall: Humans have two copies of each gene. Therefore, each human has two copies of the gene that codes for hemoglobin.
- People with two normal hemoglobin genes do not have sickle cell disease, however are susceptible to malaria.
- People with one normal and one mutant, sickle hemoglobin have sickle cell trait. This means they have some normal red blood cells and some sickle red blood cells. This allows for malaria resistance, without developing the associated disease.
- People with two mutant, sickle hemoglobin genes develop sickle cell disease. All their blood cells are sickle shaped, however they have the highest resistance to malaria.
The resistance in individuals with one or two mutant, sickle genes is due to the insufficient quantities of Potassium found in sickle cells. This causes Plasmodium to die. (Other mechanisms are still being researched)
Sickle cell disease – biointeractive video
3.1.U6 – The genome is the whole of the genetic information of an organism
Genomes and the Human Genome Project
While we have made considerable progress in the past few decades, our maps of the genomes are still far from complete.
Think about the maps of cartographers in the Middle Ages – many maps has locations that were uncharted so called Terra incognita, or unknown land. This work as an analogy for to our genetic maps in present day.
Genome – The complete set of an organisms base sequences (~3 000 000 000 in humans)
DNA sequencing:
- Sequencing is the process of determining the precise order of nucleotides with a DNA molecule.
- The complete genome of certain organisms have been sequenced (Drosophilia melanogaster, E. coli)
- The first methods for sequencing DNA were developed in the mid-1970s. However, the technology could only sequence a few base pairs a year (not even enough for a gene, let alone an entire genome).
- By 1990, a few laboratories had managed to sequence 100,000 bases, but the cost was high.
- Since then, technological improvements and automation have increased the speed and lowered the cost to the point where individual genes can be sequences routinely, and some labs can sequence well over 100 million bases per year.
Homework response: Describe the Sanger method for DNA sequencing and its limitations.
3.1.U7 – The entire base sequence of human genes was sequenced in the Human Genome Project.
The Human Genome Project
Started in 1990, the HGP had three key principles
The three principles were:
- Automatic release of sequence assemblies larger than 1kb (preferably within 24 hours).
- Immediate publication of finished annotated sequences.
- Aim to make the entire sequence freely available in the public domain for both research and development in order to maximise benefits to society.
Goal: Sequence the order of all bases in the human genome. This goal was achieved in 2003.
- The entire sequenced genome can be thought of as a map that can show the locus of any gene on any one of the 23 pairs of chromosomes.
- However, scientists are currently working on the which sequence represent genes (i.e. which are genes, which are not, which gene does what, etc.)
https://www.youtube.com/watch?v=AhsIF-cmoQQ
The HGP confirmed two (beautiful) things:
- We are all different
- We are all the same
- Remember, we share 99.9% of DNA with any given human
- However, since the genome is 3 000 000 000 base pairs long, that means there are up to 3 000 000 single base differences in each of us.
- These differences define who you are phenotypically and genotypically. But, we all share much of the same DNA

Comments by shaun pletsch